Gene Page: SLC6A12
Summary ?
| GeneID | 6539 |
| Symbol | SLC6A12 |
| Synonyms | BGT-1|BGT1|GAT2 |
| Description | solute carrier family 6 member 12 |
| Reference | MIM:603080|HGNC:HGNC:11045|Ensembl:ENSG00000111181|HPRD:04358|Vega:OTTHUMG00000090309 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.874 |
| Sherlock p-value | 0.301 |
| Fetal beta | -1.452 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3773131 | chr3 | 127429977 | SLC6A12 | 6539 | 0.11 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
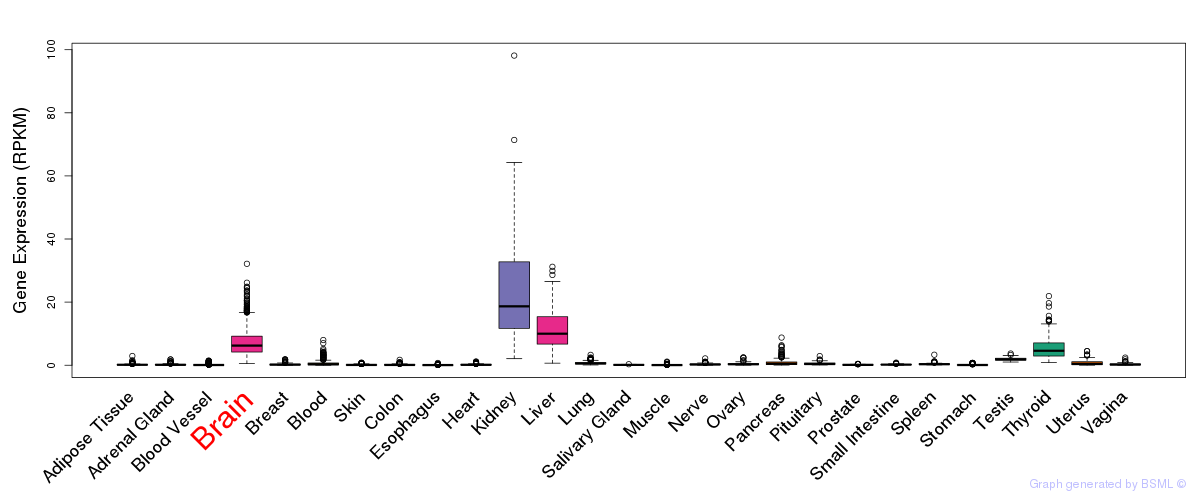
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYNM | 0.84 | 0.84 |
| AMPD3 | 0.81 | 0.80 |
| CCND1 | 0.81 | 0.86 |
| TMEM184B | 0.80 | 0.84 |
| SEPT8 | 0.80 | 0.80 |
| BCL2L2 | 0.80 | 0.79 |
| AC004690.2 | 0.80 | 0.81 |
| TOM1L2 | 0.80 | 0.81 |
| TNS1 | 0.79 | 0.83 |
| AATK | 0.78 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EXOSC8 | -0.62 | -0.66 |
| RPL31 | -0.58 | -0.70 |
| RPS23 | -0.58 | -0.70 |
| RPS13P2 | -0.57 | -0.68 |
| C9orf46 | -0.57 | -0.65 |
| RPL24 | -0.57 | -0.69 |
| SNHG12 | -0.57 | -0.67 |
| PFDN5 | -0.56 | -0.64 |
| RPL35 | -0.56 | -0.66 |
| RPS3AP47 | -0.56 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005332 | gamma-aminobutyric acid:sodium symporter activity | IEA | GABA, Neurotransmitter (GO term level: 11) | - |
| GO:0005328 | neurotransmitter:sodium symporter activity | IEA | Neurotransmitter (GO term level: 9) | - |
| GO:0015293 | symporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006836 | neurotransmitter transport | IEA | neuron, Neurotransmitter (GO term level: 5) | - |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | 31 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | 94 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | 49 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE COMPOUND SLC TRANSPORTERS | 27 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | 17 | 16 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |