Gene Page: SPRR3
Summary ?
| GeneID | 6707 |
| Symbol | SPRR3 |
| Synonyms | - |
| Description | small proline rich protein 3 |
| Reference | MIM:182271|HGNC:HGNC:11268|Ensembl:ENSG00000163209|HPRD:01652|Vega:OTTHUMG00000013872 |
| Gene type | protein-coding |
| Map location | 1q21-q22 |
| Fetal beta | 0.097 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21543102 | 1 | 152974771 | SPRR3 | 2.762E-4 | -0.298 | 0.038 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6902183 | chr6 | 121963786 | SPRR3 | 6707 | 0.1 | trans |
Section II. Transcriptome annotation
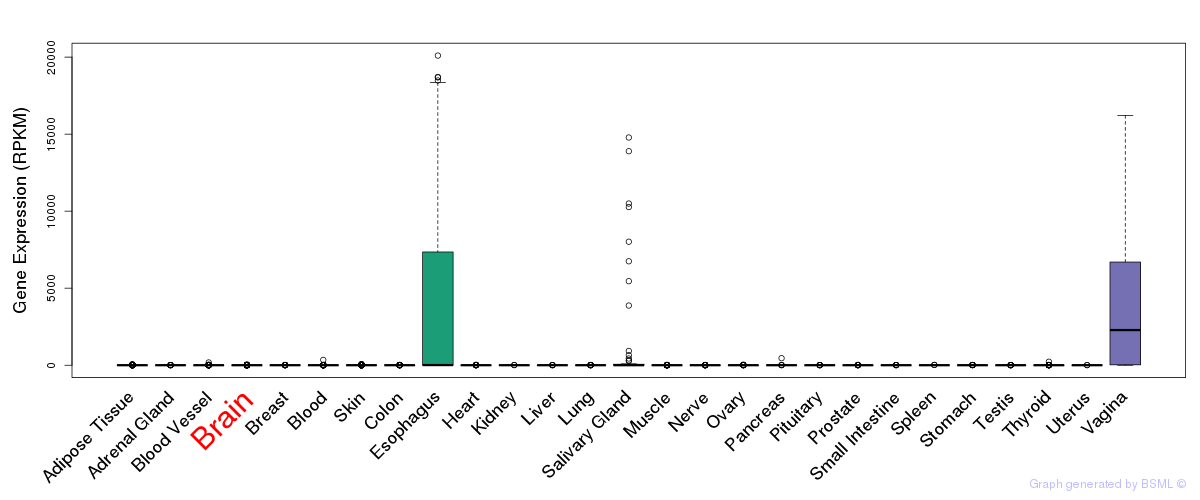
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CNIH | 0.75 | 0.72 |
| GLRX3 | 0.73 | 0.72 |
| STARD3NL | 0.71 | 0.72 |
| FAM103A1 | 0.70 | 0.68 |
| MRPL15 | 0.70 | 0.65 |
| SUCLG1 | 0.70 | 0.66 |
| BBS4 | 0.70 | 0.74 |
| ARMC10 | 0.70 | 0.70 |
| MRPS35 | 0.70 | 0.74 |
| C20orf111 | 0.69 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.45 | -0.52 |
| MT-ATP8 | -0.44 | -0.50 |
| AF347015.18 | -0.44 | -0.55 |
| AC100783.1 | -0.43 | -0.56 |
| C10orf108 | -0.43 | -0.48 |
| AF347015.15 | -0.43 | -0.50 |
| AF347015.26 | -0.43 | -0.50 |
| AF347015.27 | -0.42 | -0.49 |
| MT-CYB | -0.42 | -0.49 |
| AC016705.1 | -0.41 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10510474 | |
| GO:0005198 | structural molecule activity | TAS | 9889002 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008544 | epidermis development | NAS | 8325635 | |
| GO:0042060 | wound healing | TAS | 10510474 | |
| GO:0030216 | keratinocyte differentiation | NAS | 8325635 | |
| GO:0031424 | keratinization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS DN | 8 | 5 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP | 109 | 68 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE | 69 | 31 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |