Gene Page: TBP
Summary ?
| GeneID | 6908 |
| Symbol | TBP |
| Synonyms | GTF2D|GTF2D1|HDL4|SCA17|TFIID |
| Description | TATA-box binding protein |
| Reference | MIM:600075|HGNC:HGNC:11588|Ensembl:ENSG00000112592|HPRD:02511|Vega:OTTHUMG00000016084 |
| Gene type | protein-coding |
| Map location | 6q27 |
| Pascal p-value | 0.215 |
| Sherlock p-value | 0.118 |
| Fetal beta | 1.038 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11786005 | 6 | 170863399 | TBP | 2.24E-9 | -0.007 | 1.73E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9570060 | chr13 | 59754134 | TBP | 6908 | 0.15 | trans |
Section II. Transcriptome annotation
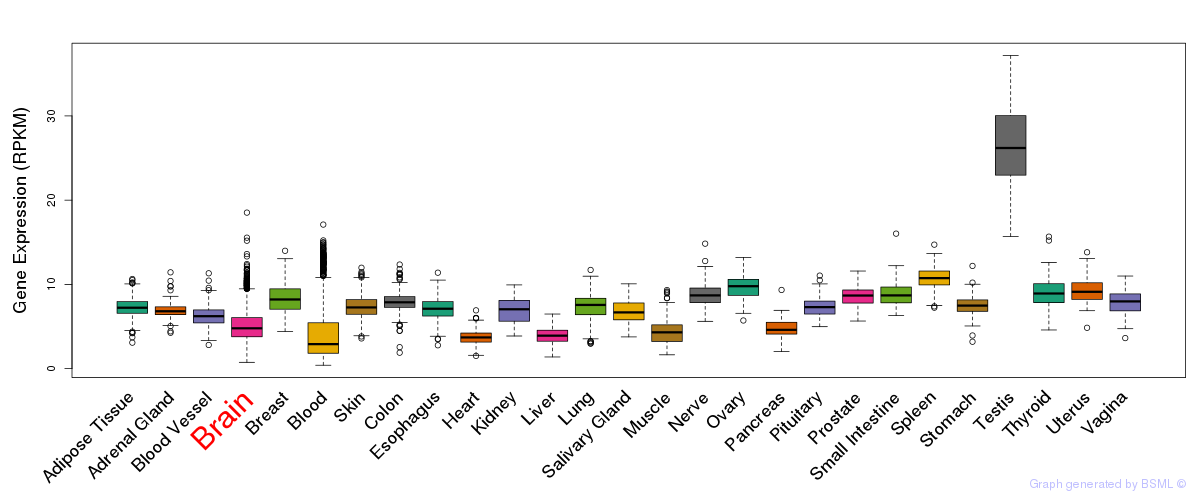
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GJA4 | 0.70 | 0.69 |
| ESAM | 0.68 | 0.69 |
| FCGRT | 0.66 | 0.68 |
| BGN | 0.65 | 0.69 |
| TIMP1 | 0.64 | 0.67 |
| MFSD2 | 0.61 | 0.64 |
| TM4SF1 | 0.61 | 0.60 |
| S100A11 | 0.61 | 0.62 |
| TGFB1 | 0.59 | 0.62 |
| ORAI3 | 0.59 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZMAT2 | -0.40 | -0.46 |
| CEP63 | -0.39 | -0.39 |
| C1orf96 | -0.38 | -0.35 |
| CIR1 | -0.36 | -0.39 |
| LMO7 | -0.36 | -0.36 |
| ZRSR2 | -0.36 | -0.37 |
| GPR22 | -0.35 | -0.38 |
| KHDRBS3 | -0.35 | -0.34 |
| PITPNB | -0.35 | -0.38 |
| RNPC3 | -0.35 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003702 | RNA polymerase II transcription factor activity | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 1465435 |7680771 |8006019 |9045704 |9748258 |10391676 |12040021 |12217962 | |
| GO:0016251 | general RNA polymerase II transcription factor activity | TAS | 2363050 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | EXP | 8946909 | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | IEA | - | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | TAS | 2363050 | |
| GO:0006368 | RNA elongation from RNA polymerase II promoter | EXP | 9405375 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008219 | cell death | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 1939271 |2449431 |8946909 | |
| GO:0005669 | transcription factor TFIID complex | IDA | 14580349 | |
| GO:0005669 | transcription factor TFIID complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABT1 | hABT1 | activator of basal transcription 1 | - | HPRD | 10648625 |
| AHR | - | aryl hydrocarbon receptor | AHR interacts with TBP. | BIND | 15641800 |
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 8794892 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Reconstituted Complex | BioGRID | 9238003 |
| ATF4 | CREB-2 | CREB2 | TAXREB67 | TXREB | activating transcription factor 4 (tax-responsive enhancer element B67) | Reconstituted Complex | BioGRID | 9295363 |
| BAX | BCL2L4 | BCL2-associated X protein | TBP interacts with the Bax promoter. | BIND | 15710329 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD,BioGRID | 9812988 |
| BRF1 | BRF | FLJ42674 | FLJ43034 | GTF3B | MGC105048 | TAF3B2 | TAF3C | TAFIII90 | TF3B90 | TFIIIB90 | hBRF | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae) | - | HPRD,BioGRID | 7624363 |10921893 |
| BRF2 | BRFU | FLJ11052 | TFIIIB50 | BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like | - | HPRD,BioGRID | 11564744 |
| BTAF1 | KIAA0940 | MGC138406 | MOT1 | TAF(II)170 | TAF172 | TAFII170 | BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae) | - | HPRD,BioGRID | 9488487 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | Affinity Capture-Western | BioGRID | 11959914 |
| CD3EAP | ASE-1 | CAST | MGC118851 | PAF49 | CD3e molecule, epsilon associated protein | Affinity Capture-Western | BioGRID | 15226435 |
| CITED2 | MRG1 | P35SRJ | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Reconstituted Complex | BioGRID | 10593900 |
| CREG1 | CREG | cellular repressor of E1A-stimulated genes 1 | - | HPRD,BioGRID | 9710587 |
| CREM | ICER | MGC111110 | MGC17881 | MGC41893 | hCREM-2 | cAMP responsive element modulator | - | HPRD,BioGRID | 10409662 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Reconstituted Complex | BioGRID | 11959865 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | - | HPRD,BioGRID | 11149922 |
| DR1 | NC2 | NC2-BETA | down-regulator of transcription 1, TBP-binding (negative cofactor 2) | - | HPRD | 11461703 |
| DRAP1 | NC2-alpha | DR1-associated protein 1 (negative cofactor 2 alpha) | - | HPRD,BioGRID | 11461703 |
| EDF1 | EDF-1 | MBF1 | MGC9058 | endothelial differentiation-related factor 1 | - | HPRD,BioGRID | 10816571 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with TBP. | BIND | 8621548 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 11595744 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | Affinity Capture-Western Reconstituted Complex | BioGRID | 8065335 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD | 7685215 |
| FOXF2 | FKHL6 | FREAC2 | forkhead box F2 | - | HPRD,BioGRID | 9722567 |
| GLI2 | HPE9 | THP1 | THP2 | GLI-Kruppel family member GLI2 | TBP interacts with GLI-2. This interaction was modeled on a demonstrated interaction between TBP from an unspecified species and human GLI-2. | BIND | 11160734 |
| GTF2A1 | MGC129969 | MGC129970 | TF2A1 | TFIIA | general transcription factor IIA, 1, 19/37kDa | Biochemical Activity Reconstituted Complex | BioGRID | 7724559 |7958900 |9603936 |11278496 |11509574 |
| GTF2A1L | ALF | MGC26254 | general transcription factor IIA, 1-like | Reconstituted Complex | BioGRID | 12107178 |
| GTF2A2 | HsT18745 | TF2A2 | TFIIA | general transcription factor IIA, 2, 12kDa | - | HPRD,BioGRID | 7724559 |7958900 |9603936 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | - | HPRD,BioGRID | 14963322 |
| GTF2E1 | FE | TF2E1 | TFIIE-A | general transcription factor IIE, polypeptide 1, alpha 56kDa | - | HPRD | 11113176 |
| GTF2E1 | FE | TF2E1 | TFIIE-A | general transcription factor IIE, polypeptide 1, alpha 56kDa | Co-purification Reconstituted Complex | BioGRID | 7926747 |9159119 |
| GTF2E2 | FE | TF2E2 | TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa | - | HPRD,BioGRID | 9677423 |11113176 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Biochemical Activity Co-purification Reconstituted Complex | BioGRID | 7590250 |9159119 |9482861 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD,BioGRID | 8577725 |
| GTF2H4 | TFB2 | TFIIH | general transcription factor IIH, polypeptide 4, 52kDa | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| GTF3C2 | KIAA0011 | TFIIIC-BETA | TFIIIC110 | general transcription factor IIIC, polypeptide 2, beta 110kDa | Reconstituted Complex | BioGRID | 10373544 |
| GTF3C3 | TFIIIC102 | TFIIICgamma | TFiiiC2-102 | general transcription factor IIIC, polypeptide 3, 102kDa | - | HPRD,BioGRID | 10373544 |
| GTF3C4 | FLJ21002 | KAT12 | MGC138450 | TFIII90 | TFIIIC90 | TFIIICdelta | TFiiiC2-90 | general transcription factor IIIC, polypeptide 4, 90kDa | Reconstituted Complex | BioGRID | 10373544 |
| GTF3C5 | FLJ20857 | TFIIIC63 | TFIIICepsilon | TFiiiC2-63 | general transcription factor IIIC, polypeptide 5, 63kDa | Reconstituted Complex | BioGRID | 10373544 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 9880542 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | - | HPRD,BioGRID | 11005381 |
| HTT | HD | IT15 | huntingtin | Htt interacts with TBP. | BIND | 15225551 |
| HTT | HD | IT15 | huntingtin | - | HPRD | 10410676 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | - | HPRD,BioGRID | 11959865 |
| IRF1 | IRF-1 | MAR | interferon regulatory factor 1 | TBP interacts with IRF-1 promoter. | BIND | 15893730 |
| JARID1A | KDM5A | RBBP2 | RBP2 | jumonji, AT rich interactive domain 1A | - | HPRD,BioGRID | 7935440 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 7848298 |
| JUND | AP-1 | jun D proto-oncogene | - | HPRD | 7848298 |
| KLF5 | BTEB2 | CKLF | IKLF | Kruppel-like factor 5 (intestinal) | Reconstituted Complex | BioGRID | 9089417 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD,BioGRID | 12392551 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Reconstituted Complex | BioGRID | 9271120 |9388200 |
| MSX1 | HOX7 | HYD1 | msh homeobox 1 | Affinity Capture-Western in vivo Phenotypic Suppression Reconstituted Complex | BioGRID | 8700832 |9111364 |10215616 |
| MSX1 | HOX7 | HYD1 | msh homeobox 1 | - | HPRD | 8700832 |10215616 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 8755740 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | TBP interacts with c-Myc. | BIND | 15723054 |
| NACA | HSD48 | MGC117224 | NACA1 | nascent polypeptide-associated complex alpha subunit | - | HPRD,BioGRID | 8835540 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 8754792 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD | 9267036 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Reconstituted Complex | BioGRID | 10567404 |
| NFYB | CBF-A | CBF-B | HAP3 | NF-YB | nuclear transcription factor Y, beta | - | HPRD,BioGRID | 9153318 |
| NFYC | CBF-C | CBFC | DKFZp667G242 | FLJ45775 | H1TF2A | HAP5 | HSM | NF-YC | nuclear transcription factor Y, gamma | Reconstituted Complex | BioGRID | 9153318 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 9649342 |
| PAX3 | CDHS | HUP2 | MGC120381 | MGC120382 | MGC120383 | MGC120384 | MGC134778 | WS1 | paired box 3 | Reconstituted Complex | BioGRID | 10359315 |
| PAX6 | AN | AN2 | D11S812E | MGC17209 | MGDA | WAGR | paired box 6 | - | HPRD,BioGRID | 10359315 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| POLR3C | RPC3 | RPC62 | polymerase (RNA) III (DNA directed) polypeptide C (62kD) | Affinity Capture-Western | BioGRID | 9171375 |
| POLR3F | MGC13517 | RPC39 | RPC6 | polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa | - | HPRD,BioGRID | 9171375 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | - | HPRD,BioGRID | 8202368 |
| POU2F2 | OCT2 | OTF2 | Oct-2 | POU class 2 homeobox 2 | - | HPRD,BioGRID | 8202368 |
| POU3F2 | BRN2 | OCT7 | OTF7 | POUF3 | POU class 3 homeobox 2 | Brn-2 interacts with TBP. | BIND | 11029584 |
| PSMC2 | MGC3004 | MSS1 | Nbla10058 | S7 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | - | HPRD,BioGRID | 11118327 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Rb interacts with TBP | BIND | 11094070 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 8413269 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 7706261 |9584164 |
| RIBIN | - | rRNA promoter binding protein | TBP interacts with rDNA. | BIND | 15723054 |
| RNF4 | RES4-26 | SNURF | ring finger protein 4 | Reconstituted Complex | BioGRID | 9710597 |
| RRN3 | DKFZp566E104 | MGC104238 | TIFIA | RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11250903 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Reconstituted Complex | BioGRID | 9843967 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 7667283 |
| SNAPC2 | PTFdelta | SNAP45 | small nuclear RNA activating complex, polypeptide 2, 45kDa | - | HPRD,BioGRID | 8633057 |
| SNAPC4 | FLJ13451 | PTFalpha | SNAP190 | small nuclear RNA activating complex, polypeptide 4, 190kDa | - | HPRD,BioGRID | 12621023 |
| SNURF | - | SNRPN upstream reading frame | Phenotypic Enhancement | BioGRID | 11696545 |
| SPIB | SPI-B | Spi-B transcription factor (Spi-1/PU.1 related) | - | HPRD,BioGRID | 10196196 |
| SUB1 | MGC102747 | P15 | PC4 | p14 | SUB1 homolog (S. cerevisiae) | - | HPRD | 9482861 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD,BioGRID | 7680771 |
| TAF10 | TAF2A | TAF2H | TAFII30 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa | - | HPRD | 7923369 |
| TAF10 | TAF2A | TAF2H | TAFII30 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa | Co-purification | BioGRID | 7680771 |9153318 |
| TAF11 | MGC:15243 | PRO2134 | TAF2I | TAFII28 | TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa | - | HPRD,BioGRID | 7729427 |
| TAF13 | MGC22425 | TAF2K | TAFII18 | TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa | - | HPRD,BioGRID | 7729427 |
| TAF15 | Npl3 | RBP56 | TAF2N | TAFII68 | hTAFII68 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 8663456 |
| TAF1A | MGC:17061 | RAFI48 | SL1 | TAFI48 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa | - | HPRD,BioGRID | 7801123 |
| TAF1A | MGC:17061 | RAFI48 | SL1 | TAFI48 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa | TBP interacts with TAFI48 within the SL1 transcription factor complex | BIND | 7801123 |
| TAF1B | MGC:9349 | RAF1B | RAFI63 | SL1 | TAFI63 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | - | HPRD,BioGRID | 7801123 |
| TAF1B | MGC:9349 | RAF1B | RAFI63 | SL1 | TAFI63 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | TBP interacts with TAFI63 within the SL1 transcription factor complex. | BIND | 7801123 |
| TAF1C | MGC:39976 | SL1 | TAFI110 | TAFI95 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa | - | HPRD,BioGRID | 7801123 |
| TAF1C | MGC:39976 | SL1 | TAFI110 | TAFI95 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa | TBP interacts with TAFI110 within the SL1 transcription factor complex | BIND | 7801123 |
| TAF1L | MGC134910 | TAF2A2 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 210kDa-like | - | HPRD,BioGRID | 12217962 |
| TAF2 | CIF150 | TAF2B | TAFII150 | TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa | TBP interacts with TAFII150. This interaction was modelled on a demonstrated interaction between human TBP and TAFII150 from Drosophila. | BIND | 7801123 |
| TAF2 | CIF150 | TAF2B | TAFII150 | TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa | Co-purification | BioGRID | 7680771 |
| TAF4 | FLJ41943 | TAF2C | TAF2C1 | TAF4A | TAFII130 | TAFII135 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | Affinity Capture-Western Co-purification | BioGRID | 9153318 |12665565 |
| TAF5 | TAF2D | TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa | - | HPRD,BioGRID | 9045704 |
| TAF6 | DKFZp781E21155 | MGC:8964 | TAF2E | TAFII70 | TAFII80 | TAFII85 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | Affinity Capture-Western Co-purification | BioGRID | 7680771 |9153318 |12665565 |
| TAF7 | TAF2F | TAFII55 | TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa | Affinity Capture-Western Co-purification | BioGRID | 9153318 |12665565 |
| TAF7L | FLJ23157 | TAF2Q | dJ738A13.1 | TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa | - | HPRD,BioGRID | 12665565 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | Co-purification | BioGRID | 9153318 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Affinity Capture-Western | BioGRID | 8650542 |
| TCEA1 | GTF2S | SII | TCEA | TF2S | TFIIS | transcription elongation factor A (SII), 1 | Reconstituted Complex | BioGRID | 9305922 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 1465435 |
| TRAM2 | KIAA0057 | translocation associated membrane protein 2 | - | HPRD | 14612417 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | - | HPRD | 10454579 |
| UBTF | NOR-90 | UBF | upstream binding transcription factor, RNA polymerase I | - | HPRD,BioGRID | 8552083 |
| WDR62 | C19orf14 | DKFZp434J046 | DKFZp686G1024 | FLJ33298 | MGC166976 | WD repeat domain 62 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASAL TRANSCRIPTION FACTORS | 36 | 24 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RARRXR PATHWAY | 15 | 9 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | 23 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 474 | 481 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-15/16/195/424/497 | 229 | 236 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-200bc/429 | 488 | 494 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-379 | 394 | 400 | 1A | hsa-miR-379brain | UGGUAGACUAUGGAACGUA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.