Gene Page: TGFB1I1
Summary ?
| GeneID | 7041 |
| Symbol | TGFB1I1 |
| Synonyms | ARA55|HIC-5|HIC5|TSC-5 |
| Description | transforming growth factor beta 1 induced transcript 1 |
| Reference | MIM:602353|HGNC:HGNC:11767|Ensembl:ENSG00000140682|HPRD:03831|Vega:OTTHUMG00000132467 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.242 |
| Fetal beta | 0.138 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25395329 | 16 | 31487852 | TGFB1I1 | 6.73E-7 | 0.361 | 0.007 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
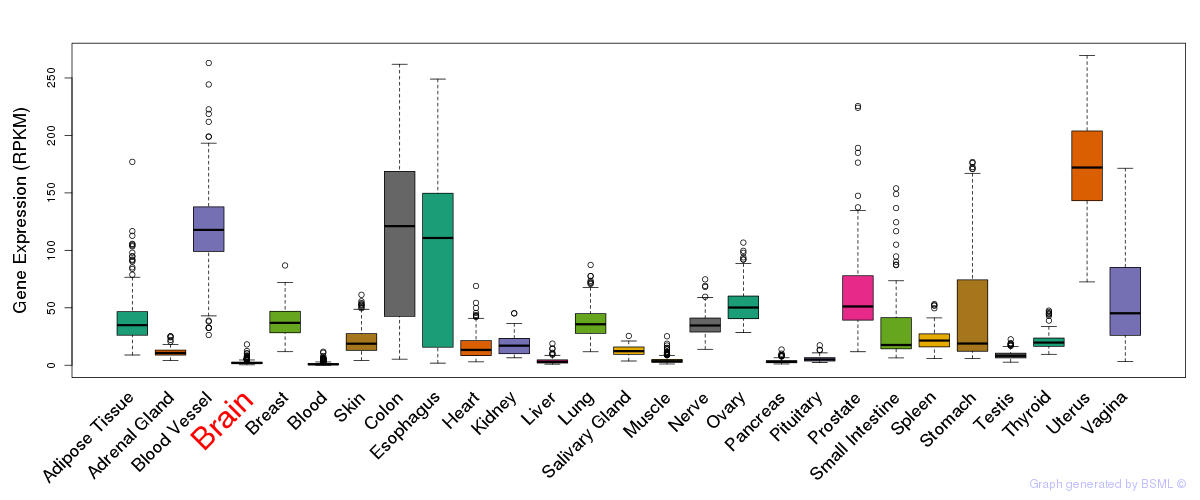
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| M6PR | 0.85 | 0.83 |
| WDR42A | 0.83 | 0.79 |
| FECH | 0.80 | 0.77 |
| HDHD2 | 0.79 | 0.75 |
| SNX9 | 0.79 | 0.78 |
| RAB5B | 0.79 | 0.74 |
| AARS | 0.79 | 0.75 |
| COG1 | 0.78 | 0.77 |
| FAM82A2 | 0.78 | 0.75 |
| APLP2 | 0.78 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IL32 | -0.53 | -0.37 |
| AF347015.21 | -0.53 | -0.25 |
| FAM159B | -0.52 | -0.53 |
| GNG11 | -0.48 | -0.34 |
| CLEC3B | -0.47 | -0.47 |
| AL050337.1 | -0.46 | -0.30 |
| AF347015.18 | -0.44 | -0.17 |
| C1orf54 | -0.44 | -0.28 |
| APOC1 | -0.43 | -0.25 |
| AC098691.2 | -0.43 | -0.31 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003713 | transcription coactivator activity | IDA | 10075738 | |
| GO:0003713 | transcription coactivator activity | NAS | 15572661 | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0050681 | androgen receptor binding | IPI | 10075738 | |
| GO:0050681 | androgen receptor binding | NAS | 15572661 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 9422762 | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 10075738 | |
| GO:0016055 | Wnt receptor signaling pathway | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 7929412 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0030521 | androgen receptor signaling pathway | NAS | 15572661 | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | NAS | 15572661 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005622 | intracellular | IDA | 10075738 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016363 | nuclear matrix | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD | 11155740 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | The carboxy terminus of ARA55 interacts with the carboxy terminus AR to enhance transcriptional activity. | BIND | 10075738 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Protein-peptide Reconstituted Complex Two-hybrid | BioGRID | 11779876 |11856738 |
| CCNA1 | - | cyclin A1 | Affinity Capture-MS | BioGRID | 17353931 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD | 9858471 |11463817 |
| CSK | MGC117393 | c-src tyrosine kinase | Reconstituted Complex | BioGRID | 9858471 |
| FKBP1A | FKBP-12 | FKBP1 | FKBP12 | FKBP12C | PKC12 | PKCI2 | PPIASE | FK506 binding protein 1A, 12kDa | - | HPRD | 12417722 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD | 12153727 |
| HSPB1 | CMT2F | DKFZp586P1322 | HMN2B | HS.76067 | HSP27 | HSP28 | Hsp25 | SRP27 | heat shock 27kDa protein 1 | Two-hybrid | BioGRID | 11546764 |
| ITGA4 | CD49D | IA4 | MGC90518 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | TGFB1I1 (Hic-5) interacts with integrin alpha-4 (ITGA4). | BIND | 10604475 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 10848625 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 9422762 |9756887 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9422762 |9858471 |11463817 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD | 9422762 |11856738 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9422762 |9858471 |11856738 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD,BioGRID | 10092676 |
| PXN | FLJ16691 | paxillin | Affinity Capture-Western | BioGRID | 11463817 |
| SDC4 | MGC22217 | SYND4 | syndecan 4 | - | HPRD | 11805099 |
| SLC6A3 | DAT | DAT1 | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | - | HPRD,BioGRID | 12177201 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | ARA55 interacts with Smad3. This interaction was modeled on a demonstrated interaction between human ARA55 and Smad3 from an unspecified species. | BIND | 15561701 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | ARA55 interacts with Smad4. This interaction was modeled on a demonstrated interaction between human ARA55 and Smad4 from an unspecified species. | BIND | 15561701 |
| SVIL | DKFZp686A17191 | supervillin | - | HPRD | 11792840 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD | 12421567 |
| TRAF4 | CART1 | MLN62 | RNF83 | TNF receptor-associated factor 4 | TRAF4 interacts with Hic-5. | BIND | 12023963 |
| VCL | CMD1W | MVCL | vinculin | Reconstituted Complex | BioGRID | 9858471 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL WORST VS BEST UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN | 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER DN | 40 | 33 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 240 MCF10A | 57 | 36 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP | 73 | 49 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL OVERALL DN | 16 | 8 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS | 32 | 22 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-199 | 102 | 108 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.