Gene Page: THRB
Summary ?
| GeneID | 7068 |
| Symbol | THRB |
| Synonyms | C-ERBA-2|C-ERBA-BETA|ERBA2|GRTH|NR1A2|PRTH|THR1|THRB1|THRB2 |
| Description | thyroid hormone receptor beta |
| Reference | MIM:190160|HGNC:HGNC:11799|Ensembl:ENSG00000151090|HPRD:07521|Vega:OTTHUMG00000130478 |
| Gene type | protein-coding |
| Map location | 3p24.2 |
| Pascal p-value | 1.81E-5 |
| Fetal beta | -1.834 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0385 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19763108 | 3 | 24187579 | THRB | 7.93E-5 | 0.596 | 0.025 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
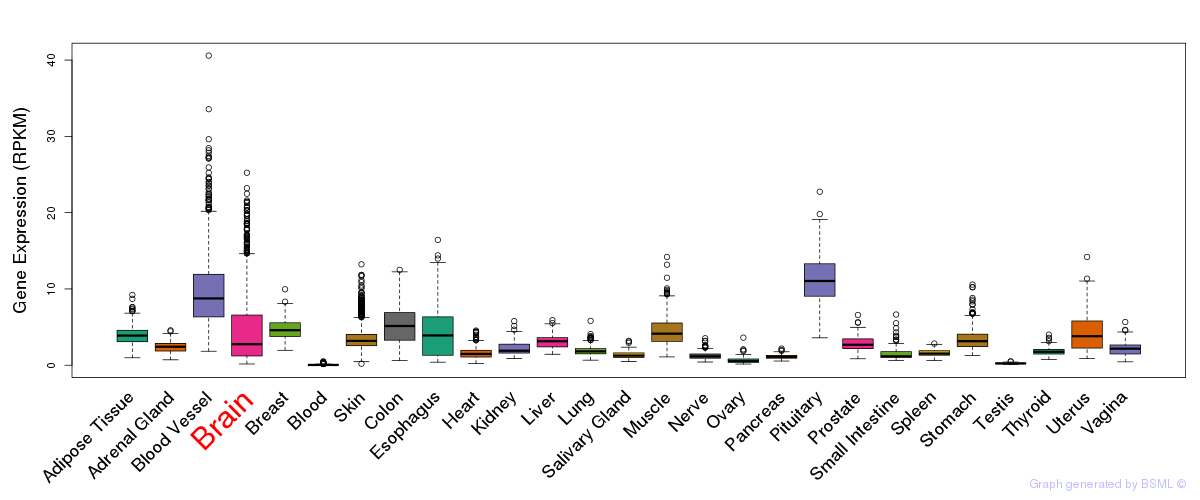
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SATB2 | 0.83 | 0.81 |
| NKAIN2 | 0.82 | 0.76 |
| TDRD7 | 0.80 | 0.79 |
| LPL | 0.79 | 0.58 |
| LGALS8 | 0.79 | 0.77 |
| SLC44A5 | 0.79 | 0.66 |
| CCBE1 | 0.79 | 0.62 |
| ADAMTSL3 | 0.78 | 0.75 |
| CHST15 | 0.77 | 0.79 |
| RBM24 | 0.77 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HSD17B14 | -0.56 | -0.61 |
| MT-CO2 | -0.53 | -0.53 |
| AF347015.31 | -0.52 | -0.51 |
| BDH2 | -0.51 | -0.55 |
| MT-CYB | -0.51 | -0.50 |
| AF347015.8 | -0.51 | -0.51 |
| AIFM3 | -0.51 | -0.50 |
| C11orf67 | -0.51 | -0.54 |
| AF347015.27 | -0.50 | -0.49 |
| AF347015.2 | -0.50 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004887 | thyroid hormone receptor activity | TAS | 1618799 | |
| GO:0003700 | transcription factor activity | NAS | 1618799 | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0003714 | transcription corepressor activity | TAS | 1618799 | |
| GO:0005515 | protein binding | IPI | 7870181 |15100213 |15625236 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0008016 | regulation of heart contraction | IEA | - | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0016481 | negative regulation of transcription | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 1618799 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANP32A | C15orf1 | I1PP2A | LANP | MAPM | MGC119787 | MGC150373 | PHAP1 | PHAPI | PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | TR-beta interacts with pp32. | BIND | 15308690 |
| BRD8 | SMAP | SMAP2 | p120 | bromodomain containing 8 | Reconstituted Complex Two-hybrid | BioGRID | 9368056 |
| BTG1 | - | B-cell translocation gene 1, anti-proliferative | BTG1 interacts with THRB (TR-beta-1). This interaction was modeled on a demonstrated interaction between human BTG1 and TR-beta-1 from an unspecified species. | BIND | 15674337 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12048199 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | - | HPRD,BioGRID | 7776974 |10207062 |
| CTCF | - | CCCTC-binding factor (zinc finger protein) | TR interacts with CTCF. | BIND | 10625678 |
| EIF3I | EIF3S2 | PRO2242 | TRIP-1 | TRIP1 | eIF3-beta | eIF3-p36 | eukaryotic translation initiation factor 3, subunit I | Two-hybrid | BioGRID | 9368056 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD | 10944117 |
| JMJD1C | DKFZp761F0118 | FLJ14374 | KIAA1380 | RP11-10C13.2 | TRIP8 | jumonji domain containing 1C | - | HPRD | 7776974 |
| MECR | CGI-63 | FASN2B | NRBF1 | mitochondrial trans-2-enoyl-CoA reductase | - | HPRD | 9795230 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD | 9325263 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | TR-beta interacts with hSrb7. | BIND | 15249124 |
| MPG | AAG | APNG | CRA36.1 | MDG | Mid1 | PIG11 | PIG16 | anpg | N-methylpurine-DNA glycosylase | MPG interacts with THRB (TR-beta). This interaction was modeled on a demonstrated interaction between MPG from an unspecified species and THRB from an unspecified species. | BIND | 14761960 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9171239 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | SRC-1 interacts with THR-beta. This interaction was modelled on a demonstrated interaction between human SRC-1 and rat TR-beta. | BIND | 9346901 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | - | HPRD,BioGRID | 11518802 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | GRIP1 interacts with TR. This interaction was modeled on a demonstrated interaction between mouse GRIP1 and human TR. | BIND | 10454563 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | TRAM-1 interacts with TR-beta. This interaction was modelled on a demonstrated interaction between human TRAM-1 and rat TR-beta. | BIND | 9346901 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | ACTR interacts with TR. | BIND | 9267036 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Reconstituted Complex Two-hybrid | BioGRID | 10567404 |10823961 |11158331 |11773444 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Reconstituted Complex | BioGRID | 9751500 |
| NR2F6 | EAR-2 | EAR2 | ERBAL2 | nuclear receptor subfamily 2, group F, member 6 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10713182 |
| NRBF2 | COPR1 | COPR2 | DKFZp564C1664 | FLJ30395 | NRBF-2 | nuclear receptor binding factor 2 | - | HPRD,BioGRID | 10786636 |
| PPARGC1A | LEM6 | PGC-1(alpha) | PGC-1v | PGC1 | PGC1A | PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | - | HPRD,BioGRID | 11751919 |
| PPARGC1B | ERRL1 | PERC | PGC-1(beta) | PGC1B | peroxisome proliferator-activated receptor gamma, coactivator 1 beta | Reconstituted Complex | BioGRID | 11733490 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | Reconstituted Complex | BioGRID | 11050077 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | PRMT2 interacts with TR-beta. | BIND | 12039952 |
| PSMC3IP | GT198 | HOP2 | HUMGT198A | TBPIP | PSMC3 interacting protein | - | HPRD | 11739747 |
| PSMC5 | S8 | SUG1 | TBP10 | TRIP1 | p45 | p45/SUG | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | Two-hybrid | BioGRID | 7870181 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Reconstituted Complex | BioGRID | 9368056 |
| RXRB | DAUDI6 | H-2RIIBP | MGC1831 | NR2B2 | RCoR-1 | retinoid X receptor, beta | THR-beta interacts with RXR-beta. This interaction was modelled on a demonstrated interaction between rat TR-beta and mouse RXR-beta. | BIND | 9346901 |
| RXRG | NR2B3 | RXRC | retinoid X receptor, gamma | - | HPRD | 9794455 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10454579 |
| TBL1X | EBI | SMAP55 | TBL1 | transducin (beta)-like 1X-linked | Reconstituted Complex | BioGRID | 10809664 |
| THRB | ERBA-BETA | ERBA2 | GRTH | MGC126109 | MGC126110 | NR1A2 | PRTH | THR1 | THRB1 | THRB2 | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | Reconstituted Complex | BioGRID | 9751500 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11258898 |
| TRIP12 | KIAA0045 | MGC138849 | MGC138850 | thyroid hormone receptor interactor 12 | - | HPRD | 7776974 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | - | HPRD,BioGRID | 7776974 |10454579 |
| TSG101 | TSG10 | VPS23 | tumor susceptibility gene 101 | Two-hybrid | BioGRID | 10508170 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGFR SMRTE PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 764 | 770 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 51 | 57 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-101 | 3770 | 3776 | m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 1472 | 1478 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA | ||||
| miR-124/506 | 1472 | 1478 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 3772 | 3778 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-135 | 1501 | 1508 | 1A,m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-138 | 280 | 286 | m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| hsa-miR-138brain | AGCUGGUGUUGUGAAUC | ||||
| miR-141/200a | 4447 | 4454 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-146 | 4321 | 4327 | m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-181 | 1410 | 1417 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-21 | 4632 | 4638 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-218 | 4167 | 4173 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-219 | 4108 | 4115 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-27 | 3772 | 3778 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-329 | 3813 | 3820 | 1A,m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-34b | 1470 | 1476 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-381 | 4543 | 4549 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-431 | 4105 | 4111 | 1A | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
| miR-452 | 4441 | 4448 | 1A,m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.