Gene Page: TNNC1
Summary ?
| GeneID | 7134 |
| Symbol | TNNC1 |
| Synonyms | CMD1Z|CMH13|TN-C|TNC|TNNC |
| Description | troponin C1, slow skeletal and cardiac type |
| Reference | MIM:191040|HGNC:HGNC:11943|Ensembl:ENSG00000114854|HPRD:08930|Vega:OTTHUMG00000158572 |
| Gene type | protein-coding |
| Map location | 3p21.1 |
| Pascal p-value | 0.036 |
| Fetal beta | -0.355 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
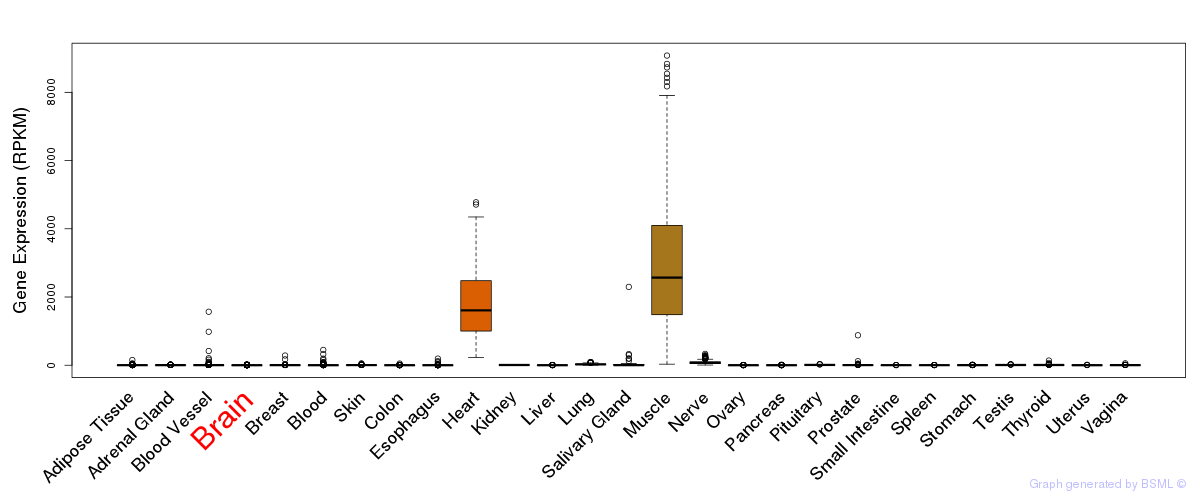
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPRML | 0.61 | 0.35 |
| GP1BB | 0.58 | 0.42 |
| NPB | 0.55 | 0.33 |
| SCARF2 | 0.52 | 0.47 |
| PRR7 | 0.52 | 0.42 |
| TFR2 | 0.51 | 0.40 |
| TNFSF9 | 0.51 | 0.37 |
| CHPF | 0.50 | 0.46 |
| TBXA2R | 0.50 | 0.33 |
| MAP1LC3A | 0.50 | 0.43 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SEC62 | -0.25 | -0.28 |
| EIF5B | -0.22 | -0.29 |
| ZNHIT6 | -0.22 | -0.20 |
| SPCS2 | -0.21 | -0.21 |
| ZNF226 | -0.21 | -0.22 |
| CHMP4A | -0.21 | -0.22 |
| CWF19L2 | -0.20 | -0.29 |
| HSPBAP1 | -0.20 | -0.23 |
| NKAPL | -0.20 | -0.21 |
| AIF1L | -0.20 | -0.11 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME STRIATED MUSCLE CONTRACTION | 27 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MUSCLE CONTRACTION | 48 | 24 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| EBAUER MYOGENIC TARGETS OF PAX3 FOXO1 FUSION | 50 | 26 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 UP | 74 | 32 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER F | 54 | 32 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 4HR | 35 | 28 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| ISSAEVA MLL2 TARGETS | 62 | 35 | All SZGR 2.0 genes in this pathway |
| WANG NFKB TARGETS | 25 | 15 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| CHEMELLO SOLEUS VS EDL MYOFIBERS UP | 35 | 16 | All SZGR 2.0 genes in this pathway |