Gene Page: UBE3A
Summary ?
| GeneID | 7337 |
| Symbol | UBE3A |
| Synonyms | ANCR|AS|E6-AP|EPVE6AP|HPVE6A |
| Description | ubiquitin protein ligase E3A |
| Reference | MIM:601623|HGNC:HGNC:12496|Ensembl:ENSG00000114062|HPRD:03375|Vega:OTTHUMG00000171548 |
| Gene type | protein-coding |
| Map location | 15q11.2 |
| Pascal p-value | 0.681 |
| Sherlock p-value | 0.63 |
| Fetal beta | 0.778 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18766912 | 15 | 25683909 | UBE3A | -0.051 | 0.29 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
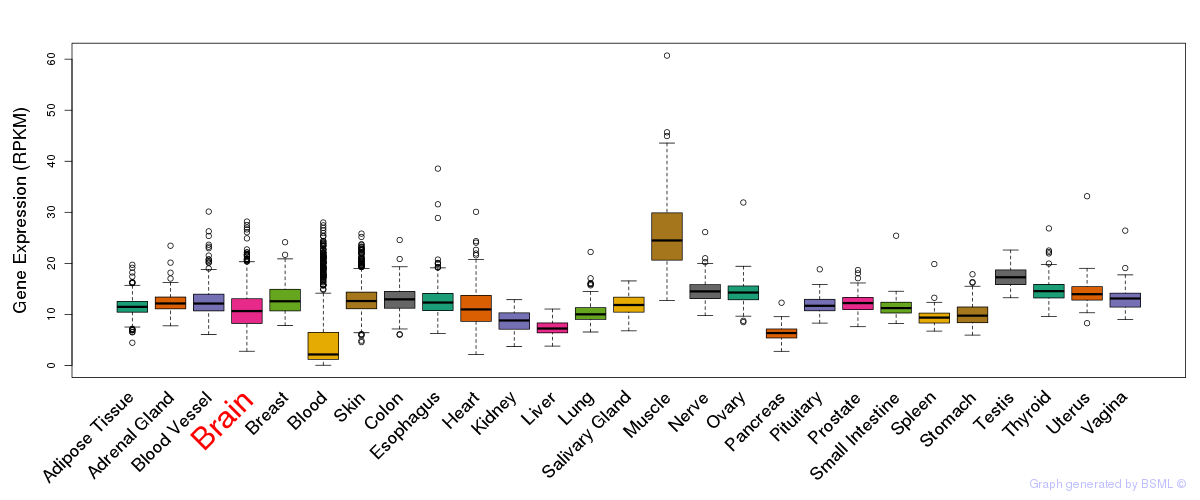
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARL2 | 0.45 | 0.30 |
| CLEC9A | 0.41 | 0.14 |
| CNFN | 0.41 | 0.22 |
| HLA-DPB1 | 0.41 | 0.29 |
| HLA-DRA | 0.39 | 0.23 |
| HLA-DMA | 0.38 | 0.22 |
| TRAC | 0.38 | 0.22 |
| HLA-DPA1 | 0.38 | 0.23 |
| CDC42EP1 | 0.38 | 0.26 |
| CD74 | 0.38 | 0.19 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GNB4 | -0.17 | -0.09 |
| DPP4 | -0.16 | -0.09 |
| C14orf37 | -0.16 | -0.08 |
| NAV3 | -0.16 | -0.06 |
| PCDH11Y | -0.16 | -0.15 |
| PDE4D | -0.16 | -0.06 |
| MPPED1 | -0.16 | -0.07 |
| SSBP2 | -0.16 | -0.04 |
| RAB39 | -0.15 | -0.05 |
| KLHL1 | -0.15 | -0.06 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16493710 | |
| GO:0004842 | ubiquitin-protein ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0016881 | acid-amino acid ligase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | TAS | Brain (GO term level: 7) | 8988171 |
| GO:0006464 | protein modification process | IEA | - | |
| GO:0006508 | proteolysis | TAS | 8221889 | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | TAS | 8221889 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0043234 | protein complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BARD1 | - | BRCA1 associated RING domain 1 | Reconstituted Complex | BioGRID | 12890688 |
| BLK | MGC10442 | B lymphoid tyrosine kinase | - | HPRD,BioGRID | 10449731 |
| BPY2 | BPY2A | VCY2 | VCY2A | basic charge, Y-linked, 2 | - | HPRD,BioGRID | 12207887 |
| BPY2B | BPY2 | VCY2 | VCY2B | basic charge, Y-linked, 2B | - | HPRD | 12207887 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Reconstituted Complex | BioGRID | 12890688 |
| CUL7 | KIAA0076 | dJ20C7.5 | cullin 7 | - | HPRD | 12481031 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | Reconstituted Complex Two-hybrid | BioGRID | 10449731 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | - | HPRD,BioGRID | 9852095 |
| PGR | NR3C3 | PR | progesterone receptor | - | HPRD,BioGRID | 9891052 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 9143503 |9369221 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Reconstituted Complex | BioGRID | 15175323 |
| UBE2D1 | E2(17)KB1 | SFT | UBC4/5 | UBCH5 | UBCH5A | ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) | - | HPRD,BioGRID | 10066826 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 9182527 |9990509 |
| UBE2D3 | E2(17)KB3 | MGC43926 | MGC5416 | UBC4/5 | UBCH5C | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | - | HPRD,BioGRID | 9990509 |
| UBE2E1 | UBCH6 | ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) | - | HPRD | 8576257 |
| UBE2G1 | E217K | UBC7 | UBE2G | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | - | HPRD | 10558980 |
| UBE2G2 | UBC7 | ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) | - | HPRD | 10558980 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | UbcH7 interacts with E6AP. | BIND | 9153201 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | Biochemical Activity Co-crystal Structure | BioGRID | 8576257 |9990509 |10558980 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD | 8576257 |10558980 |
| UBE2L6 | MGC40331 | RIG-B | UBCH8 | ubiquitin-conjugating enzyme E2L 6 | UbcH8 interacts with E6AP. | BIND | 9153201 |
| UBE2L6 | MGC40331 | RIG-B | UBCH8 | ubiquitin-conjugating enzyme E2L 6 | - | HPRD | 10558980 |
| UBQLN2 | CHAP1 | CHAP1/DSK2 | Dsk2 | HRIHFB2157 | LIC-2 | N4BP4 | PLIC-2 | PLIC2 | RIHFB2157 | ubiquilin 2 | - | HPRD,BioGRID | 10983987 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PROTEASOME PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| WONG PROTEASOME GENE MODULE | 49 | 35 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MATZUK EARLY ANTRAL FOLLICLE | 13 | 11 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP | 55 | 41 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |