Gene Page: UBTF
Summary ?
| GeneID | 7343 |
| Symbol | UBTF |
| Synonyms | NOR-90|UBF|UBF-1|UBF1|UBF2 |
| Description | upstream binding transcription factor, RNA polymerase I |
| Reference | MIM:600673|HGNC:HGNC:12511|Ensembl:ENSG00000108312|HPRD:02814|Vega:OTTHUMG00000167585 |
| Gene type | protein-coding |
| Map location | 17q21.31 |
| Pascal p-value | 0.024 |
| Sherlock p-value | 0.443 |
| Fetal beta | -0.31 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17668069 | 17 | 42295833 | UBTF | 9.67E-8 | -0.013 | 2.16E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4491879 | chr3 | 189373908 | UBTF | 7343 | 0.12 | trans |
Section II. Transcriptome annotation
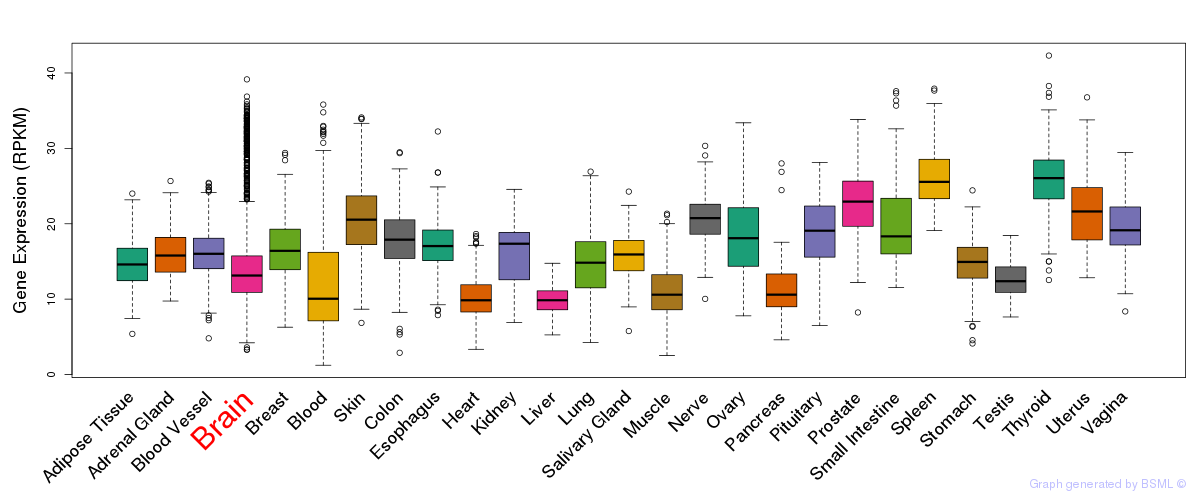
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFAB1 | 0.92 | 0.91 |
| ATP5G3 | 0.90 | 0.87 |
| COX5A | 0.89 | 0.88 |
| ATP6V0B | 0.89 | 0.89 |
| C20orf24 | 0.89 | 0.87 |
| MGST3 | 0.89 | 0.84 |
| SDHB | 0.88 | 0.89 |
| HINT1 | 0.88 | 0.81 |
| TMEM70 | 0.88 | 0.86 |
| KHDC1 | 0.87 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MTSS1L | -0.47 | -0.45 |
| GP1BA | -0.46 | -0.49 |
| AC010300.1 | -0.46 | -0.59 |
| AF347015.18 | -0.46 | -0.33 |
| C10orf108 | -0.45 | -0.46 |
| EIF5B | -0.44 | -0.50 |
| CASKIN2 | -0.44 | -0.42 |
| AC010618.2 | -0.43 | -0.45 |
| TENC1 | -0.42 | -0.44 |
| AP000769.2 | -0.42 | -0.48 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCNA2 | CCN1 | CCNA | cyclin A2 | - | HPRD | 11698641 |
| CD3EAP | ASE-1 | CAST | MGC118851 | PAF49 | CD3e molecule, epsilon associated protein | - | HPRD,BioGRID | 9426281 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | - | HPRD | 11698641 |
| CENPC1 | CENP-C | CENPC | MIF2 | hcp-4 | centromere protein C 1 | - | HPRD,BioGRID | 8702533 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 7651819 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 12554758 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD,BioGRID | 12554758 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD,BioGRID | 12748295 |
| POLR1E | FLJ13390 | FLJ13970 | PAF53 | PRAF1 | RP11-405L18.3 | polymerase (RNA) I polypeptide E, 53kDa | - | HPRD,BioGRID | 8641287 |15226435 |
| POLR1E | FLJ13390 | FLJ13970 | PAF53 | PRAF1 | RP11-405L18.3 | polymerase (RNA) I polypeptide E, 53kDa | - | HPRD | 11698641 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 11042686 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | - | HPRD | 11042686 |
| RIBIN | - | rRNA promoter binding protein | UBF interacts with rDNA. | BIND | 15723054 |
| RRN3 | DKFZp566E104 | MGC104238 | TIFIA | RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11250903 |
| SERPINH1 | AsTP3 | CBP1 | CBP2 | HSP47 | PIG14 | PPROM | RA-A47 | SERPINH2 | gp46 | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | - | HPRD | 11106745 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD,BioGRID | 12498690 |
| TAF1A | MGC:17061 | RAFI48 | SL1 | TAFI48 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa | Reconstituted Complex | BioGRID | 10913176 |
| TAF1B | MGC:9349 | RAF1B | RAFI63 | SL1 | TAFI63 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | Reconstituted Complex | BioGRID | 10913176 |
| TAF1C | MGC:39976 | SL1 | TAFI110 | TAFI95 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa | Reconstituted Complex | BioGRID | 10913176 |11698641 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 8552083 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION TERMINATION | 22 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I PROMOTER OPENING | 62 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D5 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE SULINDAC 4 | 21 | 12 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR DN | 41 | 17 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |