Gene Page: WNT3
Summary ?
| GeneID | 7473 |
| Symbol | WNT3 |
| Synonyms | INT4|TETAMS |
| Description | wingless-type MMTV integration site family member 3 |
| Reference | MIM:165330|HGNC:HGNC:12782|Ensembl:ENSG00000108379|HPRD:01318|Vega:OTTHUMG00000178076 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 6.742E-4 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17029291 | chr3 | 32402138 | WNT3 | 7473 | 0.13 | trans | ||
| rs3933652 | 17 | 44888715 | WNT3 | ENSG00000108379.5 | 4.339E-6 | 0 | 21709 | gtex_brain_ba24 |
| rs3851781 | 17 | 44891301 | WNT3 | ENSG00000108379.5 | 1.044E-6 | 0 | 19123 | gtex_brain_ba24 |
| rs77768380 | 17 | 44891767 | WNT3 | ENSG00000108379.5 | 1.429E-6 | 0 | 18657 | gtex_brain_ba24 |
| rs34724124 | 17 | 44902516 | WNT3 | ENSG00000108379.5 | 3.931E-6 | 0 | 7908 | gtex_brain_ba24 |
| rs11079738 | 17 | 44904049 | WNT3 | ENSG00000108379.5 | 1.656E-6 | 0 | 6375 | gtex_brain_ba24 |
| rs9904865 | 17 | 44908263 | WNT3 | ENSG00000108379.5 | 2.485E-17 | 0 | 2161 | gtex_brain_ba24 |
| rs2165846 | 17 | 44941366 | WNT3 | ENSG00000108379.5 | 2.215E-6 | 0 | -30942 | gtex_brain_ba24 |
| rs60372268 | 17 | 44947756 | WNT3 | ENSG00000108379.5 | 5.087E-6 | 0 | -37332 | gtex_brain_ba24 |
| rs61619946 | 17 | 44947757 | WNT3 | ENSG00000108379.5 | 5.095E-6 | 0 | -37333 | gtex_brain_ba24 |
| rs57986961 | 17 | 44947821 | WNT3 | ENSG00000108379.5 | 4.932E-6 | 0 | -37397 | gtex_brain_ba24 |
Section II. Transcriptome annotation
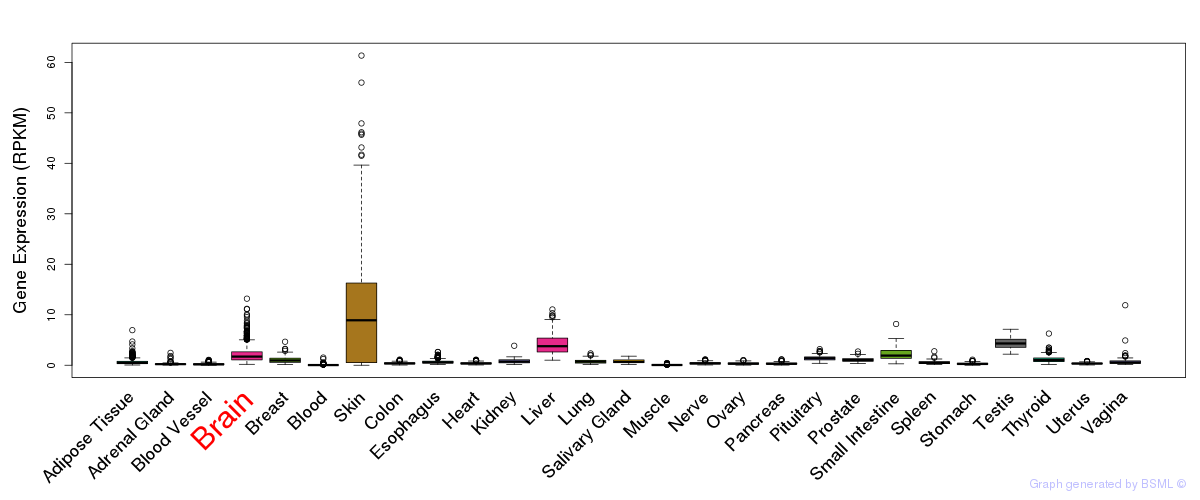
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRR11 | 0.89 | 0.62 |
| HJURP | 0.88 | 0.43 |
| PRC1 | 0.88 | 0.63 |
| GSG2 | 0.88 | 0.55 |
| C15orf42 | 0.88 | 0.56 |
| TPX2 | 0.88 | 0.56 |
| CENPF | 0.88 | 0.58 |
| HELLS | 0.88 | 0.51 |
| KIF2C | 0.87 | 0.50 |
| NCAPH | 0.87 | 0.54 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.36 | -0.52 |
| PTH1R | -0.35 | -0.43 |
| IFI27 | -0.35 | -0.52 |
| AF347015.27 | -0.35 | -0.49 |
| MT-CO2 | -0.34 | -0.50 |
| MT-CYB | -0.34 | -0.49 |
| AIFM3 | -0.34 | -0.41 |
| HLA-F | -0.34 | -0.39 |
| FBXO2 | -0.34 | -0.37 |
| SLC9A3R2 | -0.34 | -0.27 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID WNT SIGNALING PATHWAY | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER UP | 23 | 20 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |