Gene Page: LRRC2
Summary ?
| GeneID | 79442 |
| Symbol | LRRC2 |
| Synonyms | - |
| Description | leucine rich repeat containing 2 |
| Reference | MIM:607180|HGNC:HGNC:14676|Ensembl:ENSG00000163827|HPRD:06213|Vega:OTTHUMG00000133479 |
| Gene type | protein-coding |
| Map location | 3p21.31 |
| Pascal p-value | 0.161 |
| Sherlock p-value | 0.008 |
| Fetal beta | -1.827 |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1520483 | chr3 | 46510208 | LRRC2 | 79442 | 0.13 | cis | ||
| rs7641783 | chr3 | 46514271 | LRRC2 | 79442 | 0.01 | cis | ||
| rs10514713 | chr3 | 46543616 | LRRC2 | 79442 | 0.03 | cis | ||
| rs9810340 | chr3 | 46584171 | LRRC2 | 79442 | 1.679E-5 | cis | ||
| rs11919651 | chr3 | 46593905 | LRRC2 | 79442 | 0.04 | cis | ||
| rs9810340 | chr3 | 46584171 | LRRC2 | 79442 | 0 | trans | ||
| rs10509340 | chr10 | 73656981 | LRRC2 | 79442 | 0.1 | trans |
Section II. Transcriptome annotation
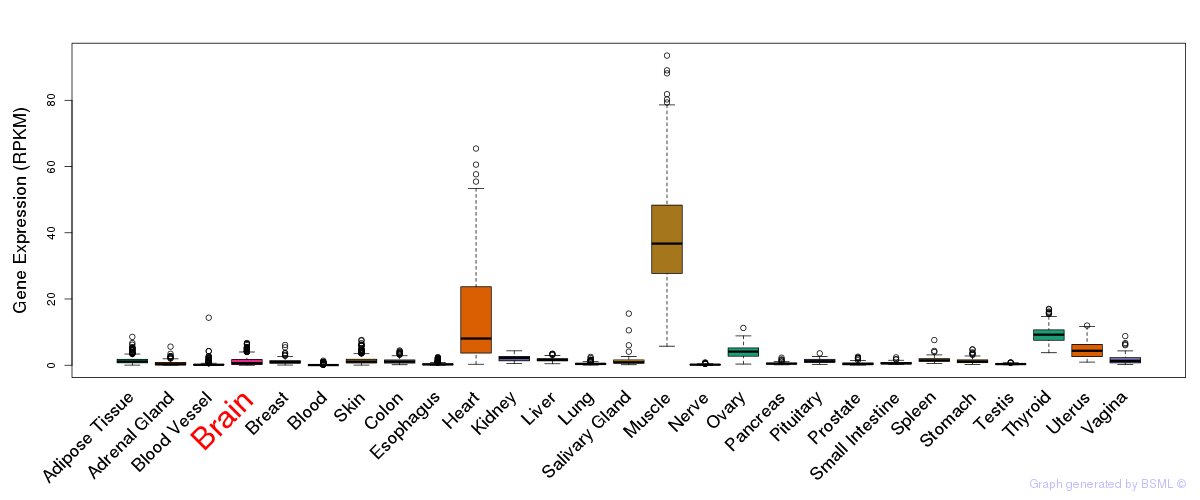
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SPAG6 | 0.91 | 0.72 |
| CASC1 | 0.90 | 0.70 |
| AC010349.1 | 0.90 | 0.76 |
| CCDC89 | 0.90 | 0.64 |
| C1orf192 | 0.90 | 0.65 |
| WDR16 | 0.90 | 0.78 |
| WDR65 | 0.90 | 0.63 |
| FHAD1 | 0.89 | 0.61 |
| C6orf165 | 0.89 | 0.57 |
| C1orf88 | 0.89 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLA | -0.26 | -0.17 |
| TMEM108 | -0.24 | -0.28 |
| DACT1 | -0.24 | -0.22 |
| SCUBE1 | -0.24 | -0.45 |
| MPPED1 | -0.24 | -0.22 |
| SH2D2A | -0.24 | -0.45 |
| VASH2 | -0.24 | -0.23 |
| AC027307.2 | -0.24 | -0.23 |
| SH3BP2 | -0.23 | -0.48 |
| ISLR2 | -0.23 | -0.20 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO METHOTREXATE UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CAMPTOTHECIN UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS UP | 112 | 65 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |