Gene Page: NUP214
Summary ?
| GeneID | 8021 |
| Symbol | NUP214 |
| Synonyms | CAIN|CAN |
| Description | nucleoporin 214kDa |
| Reference | MIM:114350|HGNC:HGNC:8064|Ensembl:ENSG00000126883|HPRD:00258|Vega:OTTHUMG00000020816 |
| Gene type | protein-coding |
| Map location | 9q34.1 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 0.951 |
| TADA p-value | 0.005 |
| Fetal beta | 0.596 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NUP214 | chr9 | 134019942 | C | NN | NM_005085 | . | frameshift | Schizophrenia | DNM:Gulsuner_2013 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09553448 | 9 | 134000424 | NUP214 | -0.057 | 0.41 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16894557 | chr6 | 28999825 | NUP214 | 8021 | 0.09 | trans | ||
| rs11975990 | chr7 | 9723034 | NUP214 | 8021 | 0.19 | trans |
Section II. Transcriptome annotation
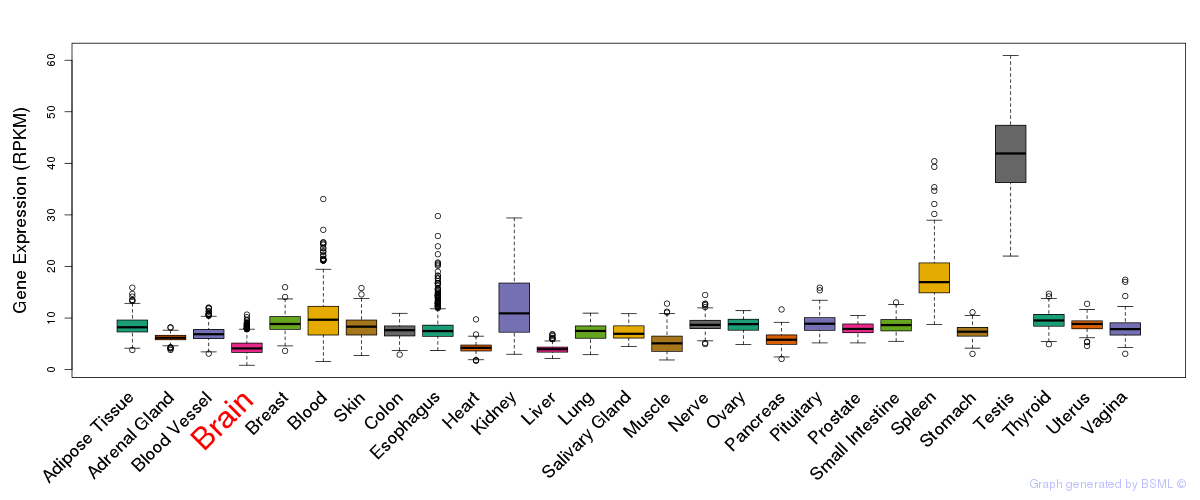
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DDX19B | DBP5 | DDX19 | RNAh | DEAD (Asp-Glu-Ala-As) box polypeptide 19B | - | HPRD,BioGRID | 10428971 |
| IPO5 | DKFZp686O1576 | FLJ43041 | IMB3 | KPNB3 | MGC2068 | RANBP5 | importin 5 | - | HPRD,BioGRID | 9114010 |
| NFX1 | DKFZp779G2416 | MGC20369 | NFX2 | nuclear transcription factor, X-box binding 1 | Reconstituted Complex | BioGRID | 11073998 |
| NUP107 | NUP84 | nucleoporin 107kDa | - | HPRD,BioGRID | 9166401 |
| NUP88 | MGC8530 | nucleoporin 88kDa | - | HPRD | 9488438 |
| NUP98 | ADIR2 | NUP196 | NUP96 | nucleoporin 98kDa | - | HPRD | 12589057 |
| NXF1 | DKFZp667O0311 | MEX67 | TAP | nuclear RNA export factor 1 | Reconstituted Complex | BioGRID | 11073998 |11551912 |
| NXF1 | DKFZp667O0311 | MEX67 | TAP | nuclear RNA export factor 1 | - | HPRD | 10668806 |11551912 |
| NXF2 | FLJ20416 | TAPL-2 | nuclear RNA export factor 2 | - | HPRD,BioGRID | 11073998 |
| NXF3 | - | nuclear RNA export factor 3 | Reconstituted Complex | BioGRID | 11073998 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 12191473 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Smad3 interacts with CAN/Nup214. This interaction was modelled on a demonstrated interaction between Smad3 and CAN/Nup214 whose origins are unknown. | BIND | 12917407 |
| TNPO1 | IPO2 | KPNB2 | MIP | MIP1 | TRN | transportin 1 | - | HPRD | 9144189 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | - | HPRD | 11551912 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | Affinity Capture-Western | BioGRID | 11425870 |
| XPO5 | FLJ14239 | FLJ32057 | FLJ45606 | KIAA1291 | exportin 5 | - | HPRD,BioGRID | 11777942 |
| XPOT | XPO3 | exportin, tRNA (nuclear export receptor for tRNAs) | - | HPRD | 12138183 |
| ZFP36 | G0S24 | GOS24 | NUP475 | RNF162A | TIS11 | TTP | zinc finger protein 36, C3H type, homolog (mouse) | - | HPRD,BioGRID | 14766228 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE TRANSPORT | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | 33 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 8 | 18 | 7 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| RAY TARGETS OF P210 BCR ABL FUSION UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |