Gene Page: BCAR3
Summary ?
| GeneID | 8412 |
| Symbol | BCAR3 |
| Synonyms | NSP2|SH2D3B |
| Description | breast cancer anti-estrogen resistance 3 |
| Reference | MIM:604704|HGNC:HGNC:973|Ensembl:ENSG00000137936|HPRD:05268|Vega:OTTHUMG00000010301 |
| Gene type | protein-coding |
| Map location | 1p22.1 |
| Pascal p-value | 0.627 |
| Sherlock p-value | 0.188 |
| Fetal beta | -0.992 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BCAR3 | chr1 | 94057915 | G | A | NM_001261408 NM_001261409 NM_001261410 NM_003567 NR_034091 | . . . . . | silent silent silent silent npcRNA | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12490747 | chr3 | 9092889 | BCAR3 | 8412 | 0.04 | trans | ||
| rs17021996 | chr4 | 147754818 | BCAR3 | 8412 | 0.13 | trans | ||
| rs17061935 | chr6 | 101873751 | BCAR3 | 8412 | 0.08 | trans | ||
| rs17080825 | chr6 | 119411222 | BCAR3 | 8412 | 0.13 | trans | ||
| rs2023879 | chr7 | 51038201 | BCAR3 | 8412 | 0.11 | trans | ||
| rs2068673 | chr12 | 60333402 | BCAR3 | 8412 | 8.293E-5 | trans | ||
| rs2655880 | chr12 | 60797980 | BCAR3 | 8412 | 0.13 | trans | ||
| rs10784054 | chr12 | 60822443 | BCAR3 | 8412 | 0.15 | trans |
Section II. Transcriptome annotation
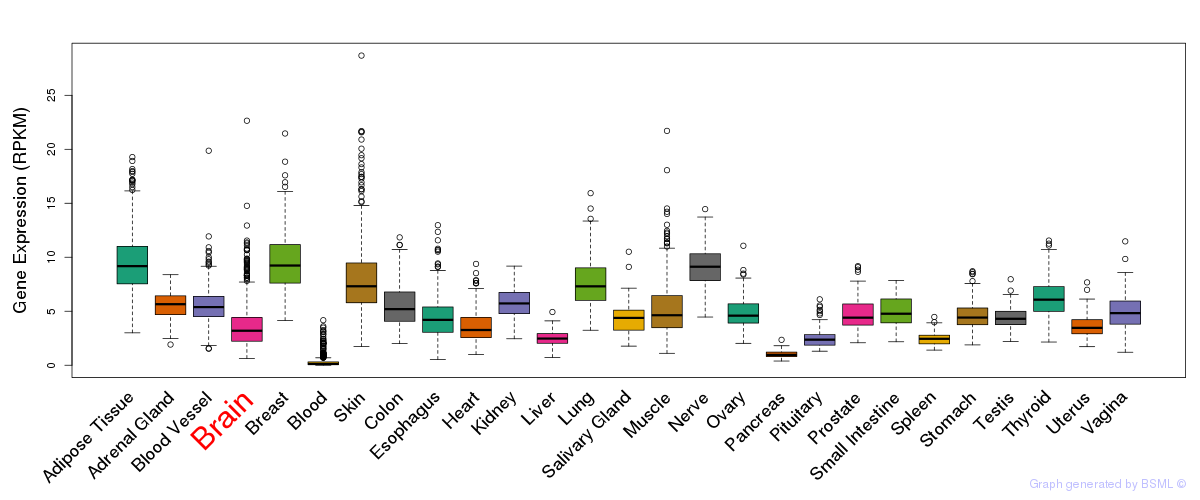
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SBNO1 | 0.93 | 0.93 |
| KIAA0232 | 0.93 | 0.94 |
| KIAA1109 | 0.92 | 0.94 |
| MYCBP2 | 0.92 | 0.92 |
| PIKFYVE | 0.91 | 0.93 |
| VPS13D | 0.91 | 0.93 |
| DMXL1 | 0.91 | 0.93 |
| KIF3A | 0.90 | 0.91 |
| ASH1L | 0.90 | 0.93 |
| KIF21A | 0.90 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf61 | -0.61 | -0.73 |
| ENHO | -0.61 | -0.66 |
| AF347015.21 | -0.60 | -0.57 |
| C1orf54 | -0.60 | -0.66 |
| RHOC | -0.59 | -0.65 |
| VAMP5 | -0.57 | -0.57 |
| METRN | -0.57 | -0.59 |
| HIGD1B | -0.57 | -0.57 |
| DBI | -0.56 | -0.62 |
| AF347015.31 | -0.56 | -0.55 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 120 MCF10A | 65 | 44 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA DN | 58 | 42 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| NOUZOVA TRETINOIN AND H4 ACETYLATION | 143 | 85 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER DN | 74 | 34 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN | 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN | 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN | 40 | 29 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |