Gene Page: CASP9
Summary ?
| GeneID | 842 |
| Symbol | CASP9 |
| Synonyms | APAF-3|APAF3|ICE-LAP6|MCH6|PPP1R56 |
| Description | caspase 9 |
| Reference | MIM:602234|HGNC:HGNC:1511|Ensembl:ENSG00000132906|HPRD:03756|Vega:OTTHUMG00000002256 |
| Gene type | protein-coding |
| Map location | 1p36.21 |
| Pascal p-value | 0.677 |
| Sherlock p-value | 0.329 |
| Fetal beta | -1.335 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20153829 | 1 | 15848307 | CASP9 | 3.371E-4 | 0.304 | 0.041 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12718316 | chr7 | 49035889 | CASP9 | 842 | 0.05 | trans | ||
| rs4132419 | chr7 | 49066632 | CASP9 | 842 | 0.16 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
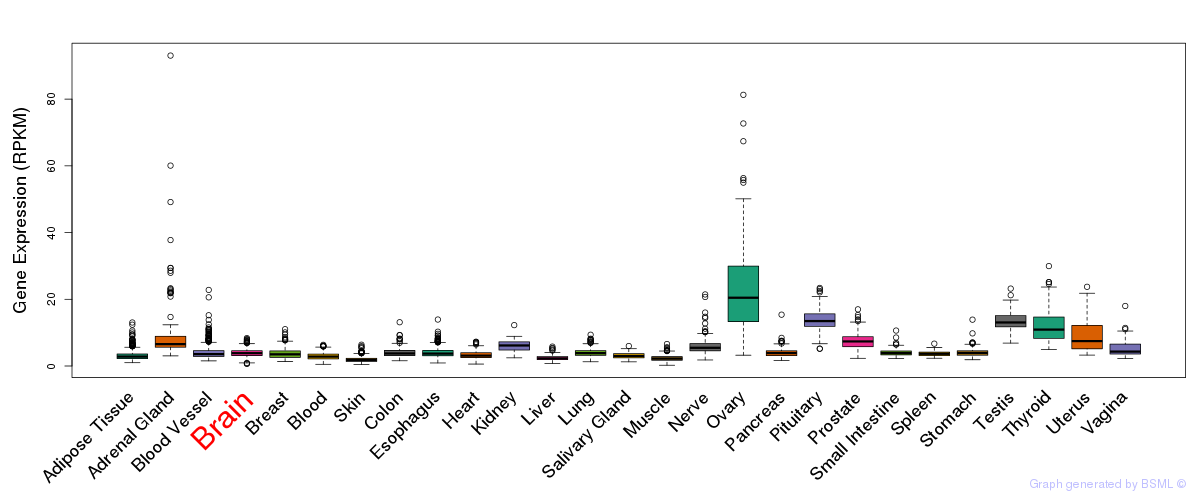
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CARD11 | 0.75 | 0.58 |
| CHRNA2 | 0.73 | 0.55 |
| ATP2A3 | 0.71 | 0.08 |
| AL133445.2 | 0.71 | 0.31 |
| SUSD2 | 0.71 | 0.38 |
| EPN3 | 0.68 | 0.43 |
| FAM19A4 | 0.68 | 0.56 |
| FAM20A | 0.68 | 0.27 |
| CHRNA3 | 0.68 | 0.44 |
| PLCB4 | 0.68 | 0.34 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PLEKHO1 | -0.18 | -0.24 |
| EMID1 | -0.17 | -0.32 |
| IER5L | -0.16 | -0.09 |
| WDR86 | -0.16 | -0.20 |
| NR2C2AP | -0.15 | -0.22 |
| TMEM44 | -0.15 | -0.18 |
| RPL35 | -0.15 | -0.30 |
| TMEM159 | -0.15 | -0.15 |
| RPL18 | -0.15 | -0.38 |
| CHRDL1 | -0.15 | -0.09 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 15657060 |
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | - | HPRD,BioGRID | 9390557 |9488720 |
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | APAF1 interacts with CASP9. | BIND | 15829969 |
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | - | HPRD | 9390557 |9488720 |9539746 |9837928 |10376594 |
| BCL10 | CARMEN | CIPER | CLAP | c-E10 | mE10 | B-cell CLL/lymphoma 10 | - | HPRD | 10187815 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | - | HPRD,BioGRID | 9539746 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | - | HPRD,BioGRID | 9545235 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | - | HPRD,BioGRID | 9545235 |
| BIRC5 | API4 | EPR-1 | baculoviral IAP repeat-containing 5 | - | HPRD,BioGRID | 11069302 |
| BIRC6 | APOLLON | BRUCE | FLJ13726 | FLJ13786 | KIAA1289 | baculoviral IAP repeat-containing 6 | - | HPRD | 15300255 |
| BIRC6 | APOLLON | BRUCE | FLJ13726 | FLJ13786 | KIAA1289 | baculoviral IAP repeat-containing 6 | BRUCE interacts with Casp-9. This interaction was modeled on a demonstrated interaction between mouse BRUCE and human Casp-9. | BIND | 15200957 |
| BIRC6 | APOLLON | BRUCE | FLJ13726 | FLJ13786 | KIAA1289 | baculoviral IAP repeat-containing 6 | Apollon interacts with and ubiquitinates Caspase-9. | BIND | 15300255 |
| BIRC7 | KIAP | LIVIN | ML-IAP | MLIAP | RNF50 | baculoviral IAP repeat-containing 7 | - | HPRD,BioGRID | 11024045 |
| BIRC8 | ILP-2 | ILP2 | hILP2 | baculoviral IAP repeat-containing 8 | - | HPRD,BioGRID | 11390657 |
| CARD8 | CARDINAL | DACAR | DKFZp779L0366 | Dakar | FLJ18119 | FLJ18121 | KIAA0955 | MGC57162 | NDPP | NDPP1 | TUCAN | caspase recruitment domain family, member 8 | - | HPRD | 11408476 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | Phenotypic Suppression | BioGRID | 11832478 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD | 11230124 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Biochemical Activity Phenotypic Suppression | BioGRID | 8962078 |11832478 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | - | HPRD | 9651578 |
| CYCS | CYC | HCS | cytochrome c, somatic | - | HPRD | 12147322 |
| DCC | CRC18 | CRCR1 | deleted in colorectal carcinoma | - | HPRD,BioGRID | 11248093 |
| DSP | DPI | DPII | desmoplakin | - | HPRD | 15500642 |
| GZMB | CCPI | CGL-1 | CGL1 | CSP-B | CSPB | CTLA1 | CTSGL1 | HLP | SECT | granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | - | HPRD | 8900201 |
| HSPD1 | CPN60 | GROEL | HSP60 | HSP65 | HuCHA60 | SPG13 | heat shock 60kDa protein 1 (chaperonin) | - | HPRD | 11230124 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD,BioGRID | 12792650 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD | 12792650 |
| MGC29506 | FLJ32987 | PACAP | hypothetical protein MGC29506 | - | HPRD | 11350957 |
| NAIP | BIRC1 | FLJ18088 | FLJ42520 | FLJ58811 | NLRB1 | psiNAIP | NLR family, apoptosis inhibitory protein | - | HPRD,BioGRID | 15280366 |
| NLRP1 | CARD7 | CLR17.1 | DEFCAP | DEFCAP-L/S | DKFZp586O1822 | KIAA0926 | NAC | NALP1 | PP1044 | SLEV1 | VAMAS1 | NLR family, pyrin domain containing 1 | - | HPRD,BioGRID | 11076957 |
| NOD1 | CARD4 | CLR7.1 | NLRC1 | nucleotide-binding oligomerization domain containing 1 | Nod1 interacts with and regulates Caspase-9 | BIND | 10329646 |
| NOD1 | CARD4 | CLR7.1 | NLRC1 | nucleotide-binding oligomerization domain containing 1 | - | HPRD,BioGRID | 10329646 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | CASP9 (caspase-9) cleaves RB-1 (hRB) at the LEND consensus site to produce the p76 form. | BIND | 15735701 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | XIAP interacts with and ubiquitinates Caspase-9. | BIND | 15300255 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | Affinity Capture-Western Far Western Reconstituted Complex Two-hybrid | BioGRID | 9545235 |11390657 |15280366 |16189514 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | - | HPRD | 9545235 |11927604 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKT PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CASPASE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA D4GDI PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MITOCHONDRIA PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HSP27 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| SA CASPASE CASCADE | 19 | 13 | All SZGR 2.0 genes in this pathway |
| SA PROGRAMMED CELL DEATH | 12 | 9 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF APOPTOSIS | 58 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE DN | 51 | 38 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA DN | 39 | 29 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB DN | 38 | 23 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| NICK RESPONSE TO PROC TREATMENT DN | 27 | 18 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |