Gene Page: STK24
Summary ?
| GeneID | 8428 |
| Symbol | STK24 |
| Synonyms | HEL-S-95|MST3|MST3B|STE20|STK3 |
| Description | serine/threonine kinase 24 |
| Reference | MIM:604984|HGNC:HGNC:11403|Ensembl:ENSG00000102572|HPRD:10385| |
| Gene type | protein-coding |
| Map location | 13q31.2-q32.3 |
| Pascal p-value | 0.454 |
| Sherlock p-value | 0.721 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04407579 | 13 | 99228509 | STK24 | 1.18E-4 | -0.199 | 0.029 | DMG:Wockner_2014 |
| cg19300307 | 13 | 99136580 | STK24 | 1.277E-4 | 0.741 | 0.03 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
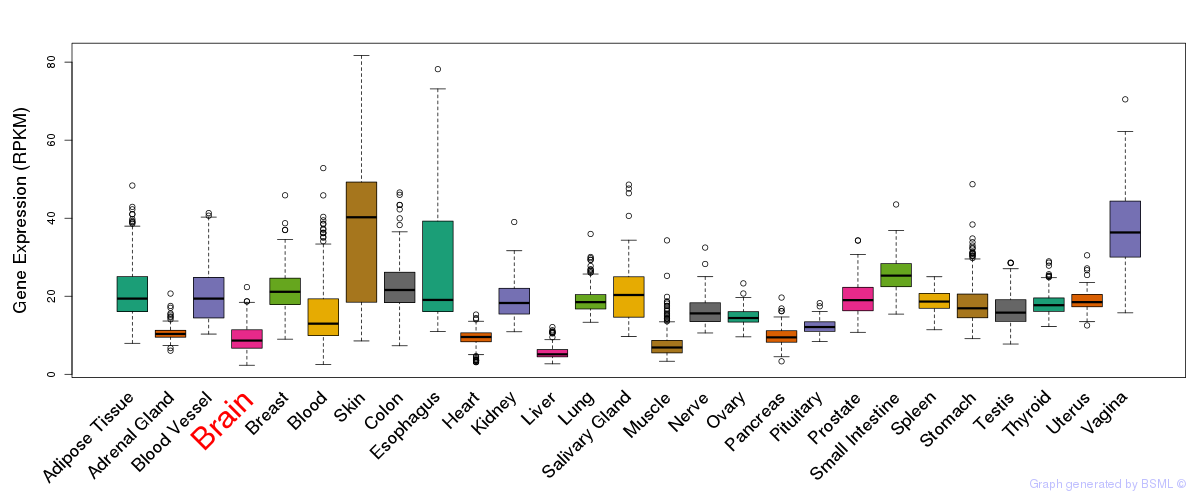
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007165 | signal transduction | TAS | 9353338 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APOD | - | apolipoprotein D | Affinity Capture-MS | BioGRID | 17353931 |
| CCT2 | 99D8.1 | CCT-beta | CCTB | MGC142074 | MGC142076 | PRO1633 | TCP-1-beta | chaperonin containing TCP1, subunit 2 (beta) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT3 | CCT-gamma | CCTG | PIG48 | TCP-1-gamma | TRIC5 | chaperonin containing TCP1, subunit 3 (gamma) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT4 | CCT-DELTA | Cctd | MGC126164 | MGC126165 | SRB | chaperonin containing TCP1, subunit 4 (delta) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT5 | CCT-epsilon | CCTE | KIAA0098 | TCP-1-epsilon | chaperonin containing TCP1, subunit 5 (epsilon) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT6A | CCT-zeta | CCT-zeta-1 | CCT6 | Cctz | HTR3 | MGC126214 | MGC126215 | MoDP-2 | TCP-1-zeta | TCP20 | TCPZ | TTCP20 | chaperonin containing TCP1, subunit 6A (zeta 1) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT7 | CCT-ETA | Ccth | MGC110985 | Nip7-1 | TCP-1-eta | chaperonin containing TCP1, subunit 7 (eta) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT8 | C21orf112 | Cctq | D21S246 | KIAA0002 | PRED71 | chaperonin containing TCP1, subunit 8 (theta) | Affinity Capture-MS | BioGRID | 18782753 |
| CTTNBP2 | C7orf8 | CORTBP2 | FLJ34229 | KIAA1758 | MGC104579 | Orf4 | cortactin binding protein 2 | Affinity Capture-MS | BioGRID | 18782753 |
| CTTNBP2NL | DKFZp547A023 | FLJ13278 | CTTNBP2 N-terminal like | Affinity Capture-MS Affinity Capture-Western | BioGRID | 18782753 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | Affinity Capture-MS | BioGRID | 17353931 |
| DSP | DPI | DPII | desmoplakin | Affinity Capture-MS | BioGRID | 17353931 |
| FABP5 | E-FABP | EFABP | PA-FABP | PAFABP | fatty acid binding protein 5 (psoriasis-associated) | Affinity Capture-MS | BioGRID | 17353931 |
| FAM40A | FLJ14743 | KIAA1761 | MGC148091 | RP4-773N10.1 | family with sequence similarity 40, member A | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| FAM40B | - | family with sequence similarity 40, member B | Affinity Capture-MS | BioGRID | 18782753 |
| FGFR1OP2 | DKFZp564O1863 | HSPC123-like | FGFR1 oncogene partner 2 | Affinity Capture-MS | BioGRID | 18782753 |
| G3BP2 | - | GTPase activating protein (SH3 domain) binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPA3 | 2610510D13Rik | D10S102 | FBRNP | HNRPA3 | MGC138232 | MGC142030 | heterogeneous nuclear ribonucleoprotein A3 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPH3 | 2H9 | FLJ34092 | HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) | Affinity Capture-MS | BioGRID | 17353931 |
| JPH3 | CAGL237 | FLJ44707 | HDL2 | JP-3 | JP3 | TNRC22 | junctophilin 3 | Affinity Capture-MS | BioGRID | 17353931 |
| MCC | DKFZp762O1615 | FLJ38893 | FLJ46755 | MCC1 | mutated in colorectal cancers | Affinity Capture-MS | BioGRID | 17353931 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| PDCD10 | CCM3 | MGC1212 | MGC24477 | TFAR15 | programmed cell death 10 | Affinity Capture-MS Two-hybrid | BioGRID | 16189514 |17353931 |18782753 |
| PDCD6IP | AIP1 | Alix | DRIP4 | HP95 | MGC17003 | programmed cell death 6 interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2CB | PP2CB | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | Affinity Capture-MS | BioGRID | 17353931 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| RP5-1000E10.4 | DKFZp686A0768 | FLJ21168 | SIKE | suppressor of IKK epsilon | Affinity Capture-MS | BioGRID | 18782753 |
| SLMAP | FLJ42206 | KIAA1601 | MGC138760 | MGC138761 | SLAP | sarcolemma associated protein | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| STK25 | DKFZp686J1430 | SOK1 | YSK1 | serine/threonine kinase 25 (STE20 homolog, yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| STRN | MGC125642 | SG2NA | striatin, calmodulin binding protein | Affinity Capture-MS Affinity Capture-Western | BioGRID | 17353931 |18782753 |
| STRN3 | SG2NA | striatin, calmodulin binding protein 3 | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| STRN4 | FLJ35594 | ZIN | zinedin | striatin, calmodulin binding protein 4 | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| SYNCRIP | GRY-RBP | HNRPQ1 | NSAP1 | RP1-3J17.2 | dJ3J17.2 | hnRNP-Q | pp68 | synaptotagmin binding, cytoplasmic RNA interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| TCP1 | CCT-alpha | CCT1 | CCTa | D6S230E | TCP-1-alpha | t-complex 1 | Affinity Capture-MS | BioGRID | 18782753 |
| TRAF3IP3 | DJ434O14.3 | FLJ44151 | MGC117354 | MGC163289 | T3JAM | TRAF3 interacting protein 3 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 18782753 |
| ZBTB24 | BIF1 | PATZ2 | ZNF450 | zinc finger and BTB domain containing 24 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK | 88 | 62 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX UP | 10 | 6 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS DN | 46 | 24 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-122 | 670 | 676 | m8 | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-128 | 482 | 488 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-139 | 509 | 515 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-190 | 793 | 799 | m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-320 | 811 | 817 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-381 | 432 | 438 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-455 | 268 | 274 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.