Gene Page: PPP1R9B
Summary ?
| GeneID | 84687 |
| Symbol | PPP1R9B |
| Synonyms | PPP1R6|PPP1R9|SPINO|Spn |
| Description | protein phosphatase 1 regulatory subunit 9B |
| Reference | MIM:603325|HGNC:HGNC:9298|Ensembl:ENSG00000108819|HPRD:04506|Vega:OTTHUMG00000162008 |
| Gene type | protein-coding |
| Map location | 17q21.33 |
| Pascal p-value | 0.287 |
| Fetal beta | 0.115 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.9202 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
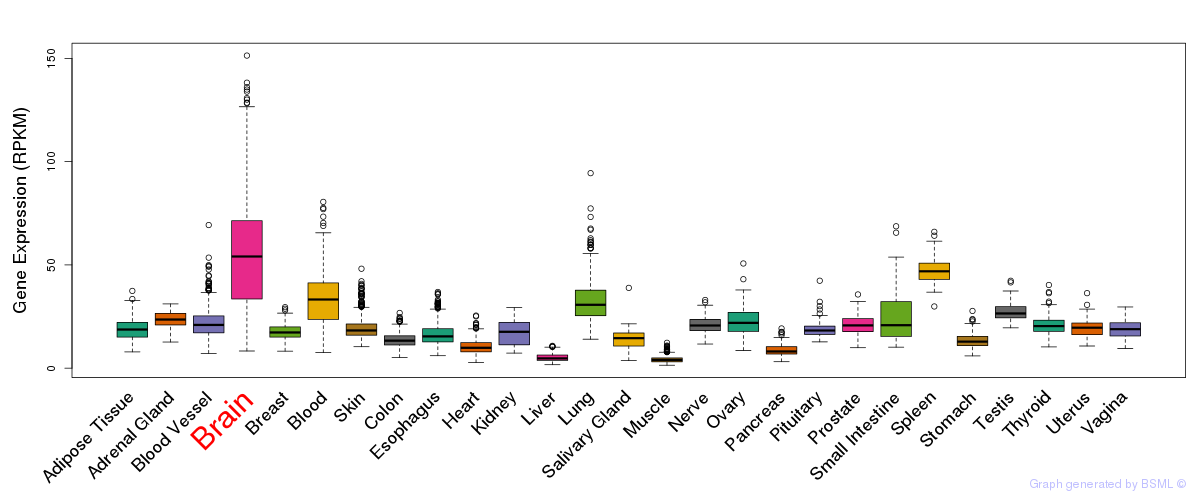
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11278317 | |
| GO:0004864 | protein phosphatase inhibitor activity | NAS | 11278317 | |
| GO:0008157 | protein phosphatase 1 binding | NAS | 11278317 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0001560 | regulation of cell growth by extracellular stimulus | TAS | 11278317 | |
| GO:0008380 | RNA splicing | NAS | 11278317 | |
| GO:0007050 | cell cycle arrest | TAS | 11278317 | |
| GO:0007096 | regulation of exit from mitosis | NAS | 11278317 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0042127 | regulation of cell proliferation | NAS | 11278317 | |
| GO:0030308 | negative regulation of cell growth | IDA | 11278317 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0000164 | protein phosphatase type 1 complex | TAS | 11278317 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | IMP | 11278317 | |
| GO:0005737 | cytoplasm | IDA | 11278317 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Reconstituted Complex | BioGRID | 12417592 |
| ACTC1 | ACTC | CMD1R | CMH11 | actin, alpha, cardiac muscle 1 | - | HPRD | 12270929|12417592 |
| ADRA1A | ADRA1C | ADRA1L1 | ALPHA1AAR | adrenergic, alpha-1A-, receptor | - | HPRD | 11154706 |
| ADRA2A | ADRA2 | ADRA2R | ADRAR | ALPHA2AAR | ZNF32 | adrenergic, alpha-2A-, receptor | Reconstituted Complex | BioGRID | 11154706 |
| ADRA2B | ADRA2L1 | ADRA2RL1 | ADRARL1 | ALPHA2BAR | adrenergic, alpha-2B-, receptor | Reconstituted Complex | BioGRID | 11154706 |
| ADRA2C | ADRA2L2 | ADRA2RL2 | ADRARL2 | ALPHA2CAR | adrenergic, alpha-2C-, receptor | Reconstituted Complex | BioGRID | 11154706 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | - | HPRD,BioGRID | 11278317 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | SPINO interacts with p14ARF. | BIND | 11278317 |
| DCX | DBCN | DC | LISX | SCLH | XLIS | doublecortin | - | HPRD | 14550532 |
| DRD2 | D2DR | D2R | dopamine receptor D2 | - | HPRD,BioGRID | 10391935 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | Affinity Capture-Western Reconstituted Complex | BioGRID | 10194355 |
| PPP1CB | MGC3672 | PP-1B | PPP1CD | protein phosphatase 1, catalytic subunit, beta isoform | Affinity Capture-Western Reconstituted Complex | BioGRID | 10194355 |
| PPP1CC | PPP1G | protein phosphatase 1, catalytic subunit, gamma isoform | Affinity Capture-Western Reconstituted Complex | BioGRID | 10194355 |10391935 |
| PPP1R2 | IPP2 | MGC87148 | protein phosphatase 1, regulatory (inhibitor) subunit 2 | - | HPRD,BioGRID | 12270929 |
| PPP1R9A | FLJ20068 | KIAA1222 | NRB1 | NRBI | Neurabin-I | protein phosphatase 1, regulatory (inhibitor) subunit 9A | - | HPRD | 10514494 |
| PPYR1 | MGC116897 | NPY4-R | NPY4R | PP1 | Y4 | pancreatic polypeptide receptor 1 | - | HPRD | 10194355 |10391935 |12270929 |
| RASGRF1 | CDC25 | CDC25L | GNRP | GRF1 | GRF55 | H-GRF55 | PP13187 | Ras protein-specific guanine nucleotide-releasing factor 1 | Affinity Capture-Western | BioGRID | 12531897 |
| TGOLN2 | MGC14722 | TGN38 | TGN46 | TGN48 | TGN51 | TTGN2 | trans-golgi network protein 2 | - | HPRD,BioGRID | 10514494 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD,BioGRID | 12531897 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO DISTAL AND PROXIMAL DENDRITES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 146 | 152 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-133 | 1143 | 1149 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU | ||||
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-148/152 | 619 | 626 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-182 | 759 | 765 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-200bc/429 | 1316 | 1322 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-216 | 440 | 446 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-335 | 431 | 437 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-34/449 | 311 | 317 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-346 | 192 | 199 | 1A,m8 | hsa-miR-346brain | UGUCUGCCCGCAUGCCUGCCUCU |
| miR-96 | 758 | 765 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.