Gene Page: LNX1
Summary ?
| GeneID | 84708 |
| Symbol | LNX1 |
| Synonyms | LNX|MPDZ|PDZRN2 |
| Description | ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| Reference | MIM:609732|HGNC:HGNC:6657|Ensembl:ENSG00000072201|HPRD:17287|Vega:OTTHUMG00000102099 |
| Gene type | protein-coding |
| Map location | 4q12 |
| Pascal p-value | 0.066 |
| Sherlock p-value | 0.777 |
| Fetal beta | -0.684 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01877606 | 4 | 54457746 | LNX1 | 1.37E-5 | -0.443 | 0.014 | DMG:Wockner_2014 |
| cg06634862 | 4 | 54457645 | LNX1 | 1.44E-5 | -0.376 | 0.014 | DMG:Wockner_2014 |
| cg23664174 | 4 | 54357316 | LNX1 | 2.232E-4 | 0.591 | 0.036 | DMG:Wockner_2014 |
| cg07080653 | 4 | 54374249 | LNX1 | 2.601E-4 | 0.655 | 0.038 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
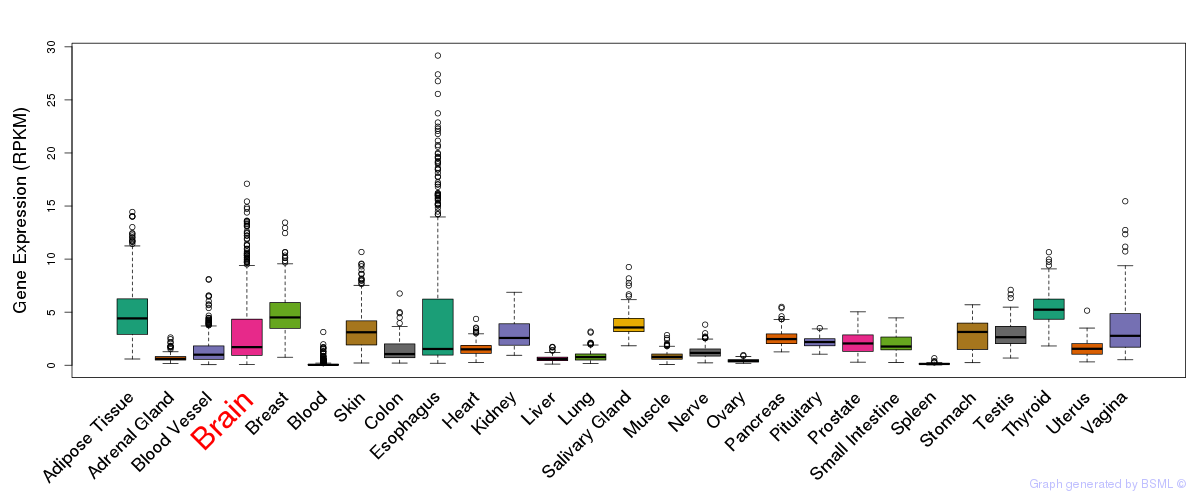
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKIRIN2 | C6orf166 | FBI1 | FLJ10342 | dJ486L4.2 | akirin 2 | Two-hybrid | BioGRID | 16189514 |
| APIP | APIP2 | CGI-29 | MMRP19 | dJ179L10.2 | APAF1 interacting protein | Two-hybrid | BioGRID | 16189514 |
| C19orf50 | FLJ25480 | MGC2749 | MST096 | MSTP096 | chromosome 19 open reading frame 50 | Two-hybrid | BioGRID | 16189514 |
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CEP72 | FLJ10565 | KIAA1519 | MGC5307 | centrosomal protein 72kDa | Two-hybrid | BioGRID | 16189514 |
| CUTC | CGI-32 | RP11-483F11.3 | cutC copper transporter homolog (E. coli) | Two-hybrid | BioGRID | 16189514 |
| DCTD | MGC111062 | dCMP deaminase | Two-hybrid | BioGRID | 16189514 |
| DDX17 | DKFZp761H2016 | P72 | RH70 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | Two-hybrid | BioGRID | 16189514 |
| DVL3 | KIAA0208 | dishevelled, dsh homolog 3 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| EHMT2 | BAT8 | C6orf30 | DKFZp686H08213 | FLJ35547 | G9A | KMT1C | NG36 | NG36/G9a | euchromatic histone-lysine N-methyltransferase 2 | Two-hybrid | BioGRID | 16189514 |
| ENOX1 | CNOX | FLJ10094 | PIG38 | bA64J21.1 | cCNOX | ecto-NOX disulfide-thiol exchanger 1 | Two-hybrid | BioGRID | 16189514 |
| FBP1 | FBP | fructose-1,6-bisphosphatase 1 | Two-hybrid | BioGRID | 16189514 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | Two-hybrid | BioGRID | 16189514 |
| JTV1 | AIMP2 | P38 | PRO0992 | JTV1 gene | Two-hybrid | BioGRID | 16189514 |
| KCTD13 | FKSG86 | PDIP1 | POLDIP1 | potassium channel tetramerisation domain containing 13 | Two-hybrid | BioGRID | 16189514 |
| KCTD17 | FLJ12242 | FLJ98761 | potassium channel tetramerisation domain containing 17 | Two-hybrid | BioGRID | 16189514 |
| KHDRBS3 | Etle | SALP | SLM-2 | SLM2 | T-STAR | TSTAR | etoile | KH domain containing, RNA binding, signal transduction associated 3 | Two-hybrid | BioGRID | 16189514 |
| KLHL12 | C3IP1 | DKIR | FLJ27152 | kelch-like 12 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| KRT15 | CK15 | K15 | K1CO | keratin 15 | Two-hybrid | BioGRID | 16189514 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| LDOC1 | BCUR1 | Mar7 | Mart7 | leucine zipper, down-regulated in cancer 1 | Two-hybrid | BioGRID | 16189514 |
| LNX2 | FLJ12933 | FLJ23932 | FLJ38000 | MGC46315 | PDZRN1 | ligand of numb-protein X 2 | - | HPRD | 11922143 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MAGEB18 | MGC33889 | melanoma antigen family B, 18 | Two-hybrid | BioGRID | 16189514 |
| NADK | FLJ13052 | FLJ37724 | dJ283E3.1 | NAD kinase | Two-hybrid | BioGRID | 16189514 |
| NAGK | GNK | HSA242910 | N-acetylglucosamine kinase | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| NUMB | S171 | numb homolog (Drosophila) | - | HPRD,BioGRID | 9535908 |11782429 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| PAICS | ADE2 | ADE2H1 | AIRC | DKFZp781N1372 | MGC1343 | MGC5024 | PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | Two-hybrid | BioGRID | 16189514 |
| PCBD1 | DCOH | PCBD | PCD | PHS | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | Two-hybrid | BioGRID | 16189514 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | Two-hybrid | BioGRID | 16189514 |
| RAD54B | FSBP | RDH54 | RAD54 homolog B (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RBMX | HNRPG | RBMXP1 | RBMXRT | RNMX | hnRNP-G | RNA binding motif protein, X-linked | Two-hybrid | BioGRID | 16189514 |
| RPIA | RPI | ribose 5-phosphate isomerase A | Two-hybrid | BioGRID | 16189514 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16189514 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | Two-hybrid | BioGRID | 16189514 |
| TIFA | MGC20791 | T2BP | T6BP | TIFAA | TRAF-interacting protein with forkhead-associated domain | Two-hybrid | BioGRID | 16189514 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| TRIM23 | ARD1 | ARFD1 | RNF46 | tripartite motif-containing 23 | Two-hybrid | BioGRID | 16189514 |
| TRIP13 | 16E1BP | thyroid hormone receptor interactor 13 | Two-hybrid | BioGRID | 16189514 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| ZBTB43 | ZBTB22B | ZNF-X | ZNF297B | zinc finger and BTB domain containing 43 | Two-hybrid | BioGRID | 16189514 |
| ZBTB8 | BOZF1 | FLJ90065 | MGC17919 | zinc finger and BTB domain containing 8 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| NEWMAN ERCC6 TARGETS DN | 39 | 24 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |