Gene Page: TANC1
Summary ?
| GeneID | 85461 |
| Symbol | TANC1 |
| Synonyms | ROLSB|TANC |
| Description | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| Reference | MIM:611397|HGNC:HGNC:29364|Ensembl:ENSG00000115183|HPRD:18146|Vega:OTTHUMG00000153945 |
| Gene type | protein-coding |
| Map location | 2q24.2 |
| Pascal p-value | 0.703 |
| Sherlock p-value | 0.429 |
| Fetal beta | -1.127 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
| Support | CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02395 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TANC1 | chr2 | 160074125 | G | A | NM_001145909 NM_033394 | p.1113R>H p.1121R>H | missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04852339 | 2 | 159841571 | TANC1 | 1.668E-4 | -0.396 | 0.033 | DMG:Wockner_2014 |
| cg06961147 | 2 | 159824263 | TANC1 | 5.074E-4 | -0.478 | 0.047 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7000604 | chr8 | 74746197 | TANC1 | 85461 | 0.1 | trans |
Section II. Transcriptome annotation
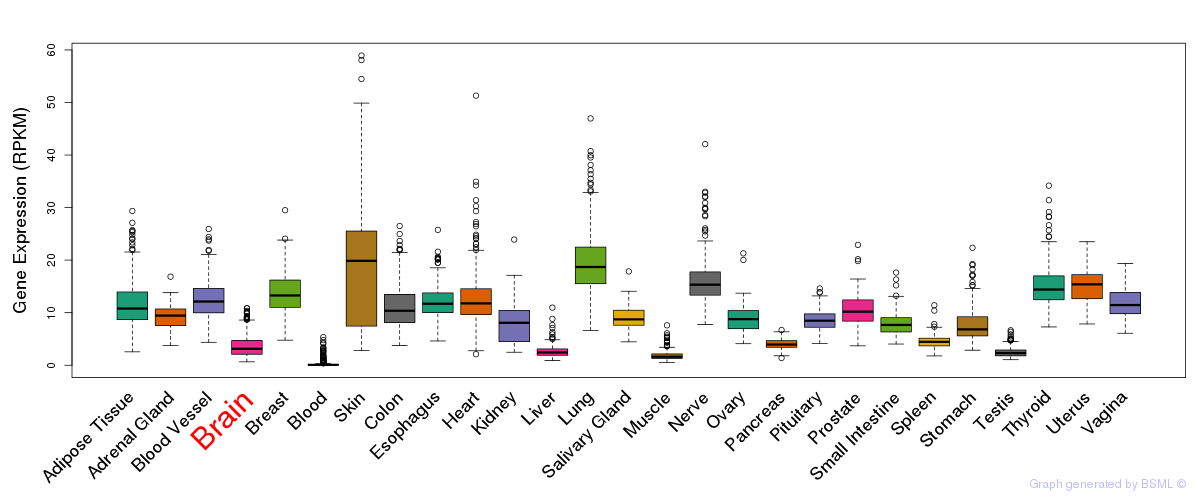
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005488 | binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 15673434 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | - | HPRD | 15673434 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD | 15673434 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD | 15673434 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD | 15673434 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | - | HPRD | 15673434 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | - | HPRD | 15673434 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD | 15673434 |
| HOMER1 | HOMER | HOMER1A | HOMER1B | HOMER1C | SYN47 | Ves-1 | homer homolog 1 (Drosophila) | - | HPRD | 15673434 |
| INA | FLJ18662 | FLJ57501 | MGC12702 | NEF5 | NF-66 | TXBP-1 | internexin neuronal intermediate filament protein, alpha | - | HPRD | 15673434 |
| SHANK1 | SPANK-1 | SSTRIP | synamon | SH3 and multiple ankyrin repeat domains 1 | - | HPRD | 15673434 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | - | HPRD | 15673434 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB UP | 20 | 12 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C DN | 59 | 39 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 DN | 17 | 13 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1608 | 1614 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-142-5p | 1122 | 1128 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 1608 | 1614 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-17-5p/20/93.mr/106/519.d | 1120 | 1127 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-23 | 1316 | 1322 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-29 | 1152 | 1158 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-338 | 1060 | 1066 | m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-543 | 1366 | 1372 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.