Gene Page: PIAS1
Summary ?
| GeneID | 8554 |
| Symbol | PIAS1 |
| Synonyms | DDXBP1|GBP|GU/RH-II|ZMIZ3 |
| Description | protein inhibitor of activated STAT 1 |
| Reference | MIM:603566|HGNC:HGNC:2752|HPRD:16029| |
| Gene type | protein-coding |
| Map location | 15q |
| Pascal p-value | 0.58 |
| Sherlock p-value | 0.148 |
| Fetal beta | 0.416 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06790275 | 15 | 68346669 | PIAS1 | -0.027 | 0.57 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
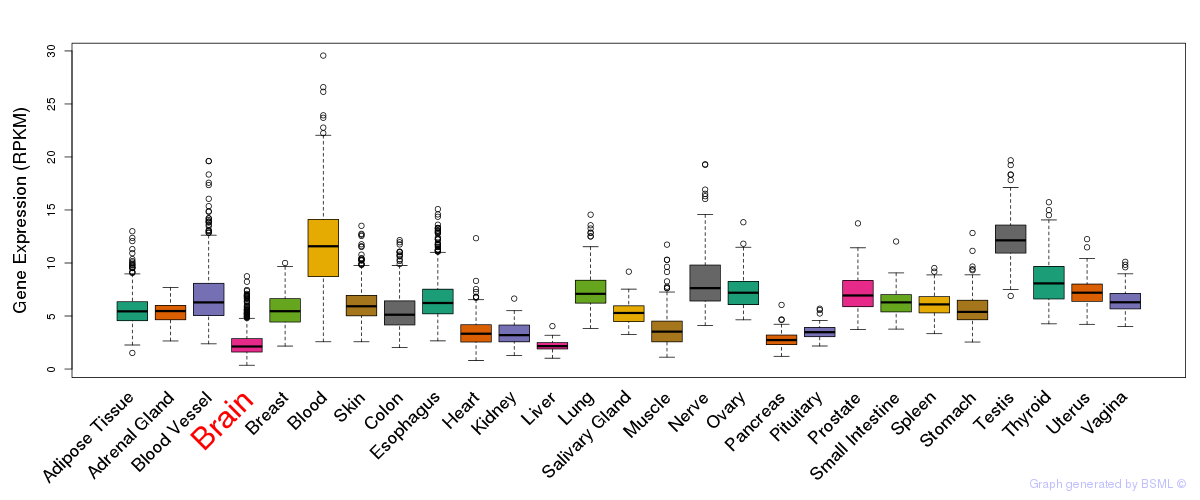
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PHF23 | 0.82 | 0.85 |
| TMED9 | 0.81 | 0.82 |
| ADRM1 | 0.79 | 0.82 |
| TIMM22 | 0.79 | 0.79 |
| CTNNBIP1 | 0.79 | 0.82 |
| R3HCC1 | 0.79 | 0.79 |
| POLR2E | 0.79 | 0.80 |
| SAMM50 | 0.79 | 0.80 |
| GRWD1 | 0.79 | 0.82 |
| VPS4A | 0.79 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.70 | -0.73 |
| AF347015.31 | -0.70 | -0.71 |
| AF347015.2 | -0.68 | -0.74 |
| AF347015.8 | -0.68 | -0.72 |
| MT-CYB | -0.67 | -0.71 |
| AF347015.33 | -0.67 | -0.70 |
| AF347015.15 | -0.66 | -0.71 |
| AF347015.27 | -0.65 | -0.70 |
| AF347015.21 | -0.64 | -0.67 |
| AF347015.9 | -0.61 | -0.69 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD | 11877418 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Two-hybrid | BioGRID | 11117529 |
| AXIN1 | AXIN | MGC52315 | axin 1 | - | HPRD | 12223491 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | CASP8 (caspase-8) interacts with PIAS-1. | BIND | 15782135 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | C/EBP-alpha interacts with PIAS1. | BIND | 15588942 |
| CEBPE | C/EBP-epsilon | CRP1 | CCAAT/enhancer binding protein (C/EBP), epsilon | C/EBP-epsilon interacts with PIAS1. | BIND | 15588942 |
| CEBPE | C/EBP-epsilon | CRP1 | CCAAT/enhancer binding protein (C/EBP), epsilon | - | HPRD | 15588942 |
| CSRP2 | CRP2 | LMO5 | SmLIM | cysteine and glycine-rich protein 2 | CRP2 interacts with PIAS1. | BIND | 11672422 |
| CSRP2 | CRP2 | LMO5 | SmLIM | cysteine and glycine-rich protein 2 | - | HPRD,BioGRID | 11672422 |
| DCLRE1A | KIAA0086 | PSO2 | SNM1 | SNM1A | hSNM1 | DNA cross-link repair 1A (PSO2 homolog, S. cerevisiae) | - | HPRD | 15572677 |
| DDX21 | DKFZp686F21172 | GUA | GURDB | RH-II/GU | RH-II/GuA | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | - | HPRD | 9177271 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 15123615 |
| DNMT3A | DNMT3A2 | M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha | - | HPRD,BioGRID | 14752048 |
| ELK3 | ERP | NET | SAP2 | ELK3, ETS-domain protein (SRF accessory protein 2) | - | HPRD | 15580297 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER-alpha interacts with PIAS1. | BIND | 15666801 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD | 15666801 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD | 11867732 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD | 12393906 |
| NR2F2 | ARP1 | COUP-TFII | COUPTFB | MGC117452 | SVP40 | TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 | - | HPRD | 15611122 |15666827 |
| NR3C2 | MCR | MGC133092 | MLR | MR | nuclear receptor subfamily 3, group C, member 2 | - | HPRD | 14500761 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | - | HPRD | 11877418 |
| PIAS4 | FLJ12419 | MGC35296 | PIASY | Piasg | ZMIZ6 | protein inhibitor of activated STAT, 4 | - | HPRD | 11877418 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 14500712 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD | 15657437 |
| SATB2 | FLJ21474 | FLJ32076 | KIAA1034 | MGC119474 | MGC119477 | SATB homeobox 2 | - | HPRD | 14701874 |
| SP100 | DKFZp686E07254 | FLJ00340 | FLJ34579 | SP100 nuclear antigen | Biochemical Activity | BioGRID | 18691969 |
| SP3 | DKFZp686O1631 | SPR-2 | Sp3 transcription factor | - | HPRD,BioGRID | 12356736 |
| STAB2 | DKFZp434E0321 | FEEL-2 | FELE-2 | FELL | FELL-2 | FEX2 | HARE | STAB-2 | stabilin 2 | Affinity Capture-Western | BioGRID | 14701874 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 9724754 |10805787 |12855578|10805787 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD | 9724754 |10805787 |12855578 |
| SUFU | PRO1280 | SUFUH | SUFUXL | suppressor of fused homolog (Drosophila) | - | HPRD | 14611647 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | SUMO-1 interacts with PIAS1. | BIND | 14609633 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD,BioGRID | 11583632 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 10380882 |11583632 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | PIAS1 interacts with p53. This interaction was modeled on a demonstrated interaction between human PIAS1 and p53 from an unspecified species. | BIND | 15133049 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 11583632 |11788578 |11867732 |15133049 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Ubc9 interacts with PIAS1. | BIND | 14609633 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD | 12177000 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Reconstituted Complex Two-hybrid | BioGRID | 11583632 |12356736 |14609633 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| ST JAK STAT PATHWAY | 9 | 8 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID RANBP2 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNG SIGNALING | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO ANDROGEN UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO FORSKOLIN DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| AMIT DELAYED EARLY GENES | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 40 HELA | 42 | 29 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |