Gene Page: EIF3G
Summary ?
| GeneID | 8666 |
| Symbol | EIF3G |
| Synonyms | EIF3-P42|EIF3S4|eIF3-delta|eIF3-p44 |
| Description | eukaryotic translation initiation factor 3 subunit G |
| Reference | MIM:603913|HGNC:HGNC:3274|Ensembl:ENSG00000130811|HPRD:04886|Vega:OTTHUMG00000180396 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.884 |
| Sherlock p-value | 0.997 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0018 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24911803 | 19 | 10230727 | EIF3G | 3.61E-8 | -0.013 | 1.05E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
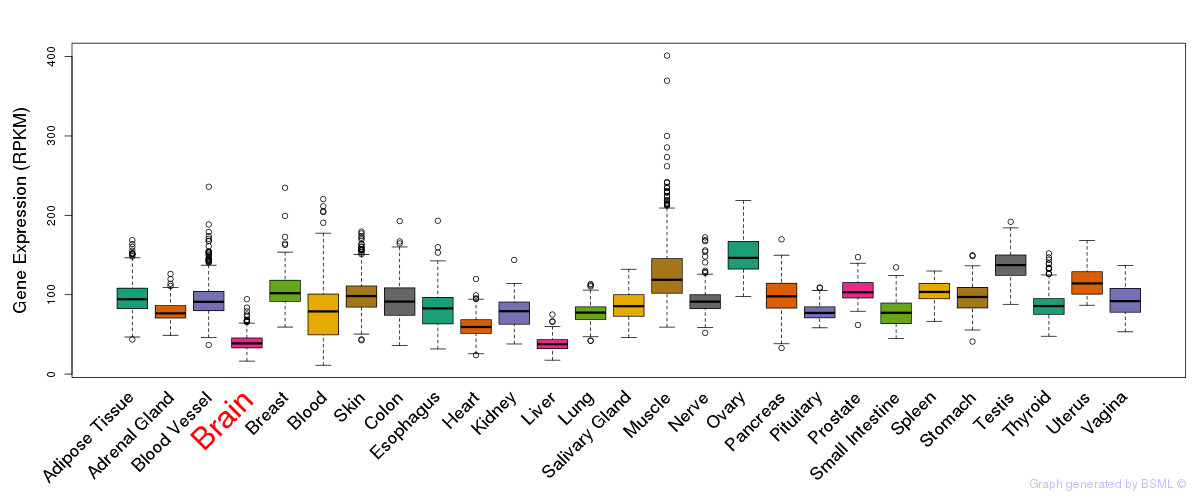
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NEK9 | 0.82 | 0.82 |
| PKD2 | 0.79 | 0.79 |
| ADAM17 | 0.78 | 0.75 |
| TBC1D2B | 0.78 | 0.78 |
| GOLGA1 | 0.78 | 0.79 |
| IKBKB | 0.77 | 0.79 |
| POFUT2 | 0.77 | 0.75 |
| CAPN2 | 0.76 | 0.79 |
| FUBP3 | 0.76 | 0.74 |
| YES1 | 0.75 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MYL3 | -0.49 | -0.50 |
| AF347015.21 | -0.49 | -0.53 |
| IFI27 | -0.47 | -0.46 |
| AF347015.31 | -0.46 | -0.49 |
| AC044839.2 | -0.46 | -0.49 |
| RERGL | -0.46 | -0.50 |
| CCDC85B | -0.46 | -0.49 |
| NENF | -0.45 | -0.53 |
| AF347015.27 | -0.45 | -0.46 |
| COX5B | -0.45 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0003743 | translation initiation factor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 14688252 |15231747 |17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006446 | regulation of translational initiation | TAS | 9973622 | |
| GO:0006412 | translation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 12588972 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | 74 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 3 | 28 | 16 | All SZGR 2.0 genes in this pathway |
| LUI TARGETS OF PAX8 PPARG FUSION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI DN | 73 | 45 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| CHNG MULTIPLE MYELOMA HYPERPLOID UP | 52 | 25 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G5 | 12 | 8 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |