Gene Page: CCNK
Summary ?
| GeneID | 8812 |
| Symbol | CCNK |
| Synonyms | CPR4 |
| Description | cyclin K |
| Reference | MIM:603544|HGNC:HGNC:1596| |
| Gene type | protein-coding |
| Map location | 14q32 |
| Pascal p-value | 0.176 |
| Sherlock p-value | 0.224 |
| Fetal beta | 0.214 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14554101 | 14 | 99948187 | CCNK | 1.48E-10 | -0.012 | 5E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
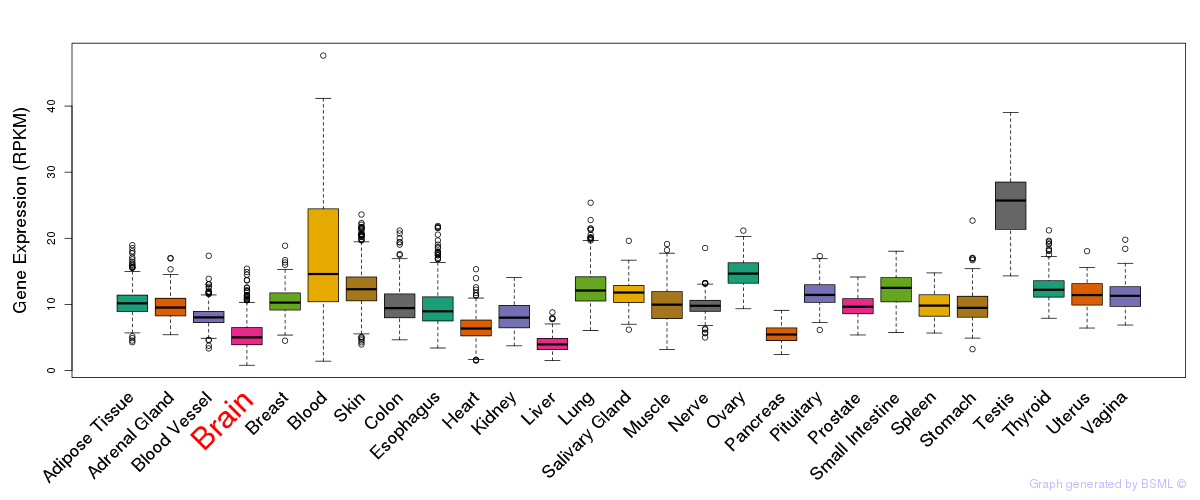
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PSMD13 | 0.91 | 0.91 |
| TRAPPC3 | 0.91 | 0.89 |
| PSMA5 | 0.91 | 0.89 |
| PSMA7 | 0.90 | 0.91 |
| LCMT1 | 0.90 | 0.88 |
| ARPC2 | 0.90 | 0.87 |
| PSMB7 | 0.90 | 0.92 |
| DCTN2 | 0.89 | 0.87 |
| CCT7 | 0.88 | 0.87 |
| C1orf50 | 0.87 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.79 | -0.79 |
| AF347015.8 | -0.79 | -0.77 |
| AF347015.15 | -0.79 | -0.79 |
| AF347015.26 | -0.78 | -0.80 |
| MT-CYB | -0.78 | -0.77 |
| MT-CO2 | -0.78 | -0.73 |
| AF347015.27 | -0.78 | -0.78 |
| AF347015.33 | -0.77 | -0.76 |
| AF347015.31 | -0.76 | -0.74 |
| MT-ATP8 | -0.70 | -0.77 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CCNT1 | CCNT | CYCT1 | cyclin T1 | in vitro in vivo | BioGRID | 10574912 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | in vitro in vivo Two-hybrid | BioGRID | 10574912 |
| CEP76 | C18orf9 | FLJ12542 | HsT1705 | centrosomal protein 76kDa | Two-hybrid | BioGRID | 16189514 |
| DVL3 | KIAA0208 | dishevelled, dsh homolog 3 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| EFEMP2 | FBLN4 | MBP1 | UPH1 | EGF-containing fibulin-like extracellular matrix protein 2 | Two-hybrid | BioGRID | 16189514 |
| FKBP6 | FKBP36 | MGC87179 | PPIase | FK506 binding protein 6, 36kDa | Two-hybrid | BioGRID | 16189514 |
| HMG20A | FLJ10739 | HMGX1 | high-mobility group 20A | Two-hybrid | BioGRID | 16189514 |
| KCNRG | DLTET | potassium channel regulator | Two-hybrid | BioGRID | 16189514 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| PCTK3 | PCTAIRE | PCTAIRE3 | PCTAIRE protein kinase 3 | Two-hybrid | BioGRID | 16189514 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western in vitro | BioGRID | 9632813 |10574912 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| RFX6 | MGC33442 | RFXDC1 | dJ955L16.1 | regulatory factor X, 6 | Two-hybrid | BioGRID | 16189514 |
| RHOXF2 | PEPP-2 | PEPP2 | THG1 | Rhox homeobox family, member 2 | Two-hybrid | BioGRID | 16189514 |
| STX11 | FHL4 | HLH4 | HPLH4 | syntaxin 11 | Two-hybrid | BioGRID | 16189514 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| USHBP1 | AIEBP | FLJ38709 | FLJ90681 | MCC2 | Usher syndrome 1C binding protein 1 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR DN | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN | 40 | 29 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |