Gene Page: TSC22D1
Summary ?
| GeneID | 8848 |
| Symbol | TSC22D1 |
| Synonyms | Ptg-2|TGFB1I4|TSC22 |
| Description | TSC22 domain family member 1 |
| Reference | MIM:607715|HGNC:HGNC:16826|Ensembl:ENSG00000102804|HPRD:07414|Vega:OTTHUMG00000016838 |
| Gene type | protein-coding |
| Map location | 13q14 |
| Pascal p-value | 0.327 |
| Sherlock p-value | 0.354 |
| Fetal beta | 0.303 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0363 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22183626 | 13 | 45150990 | TSC22D1 | -0.03 | 0.25 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
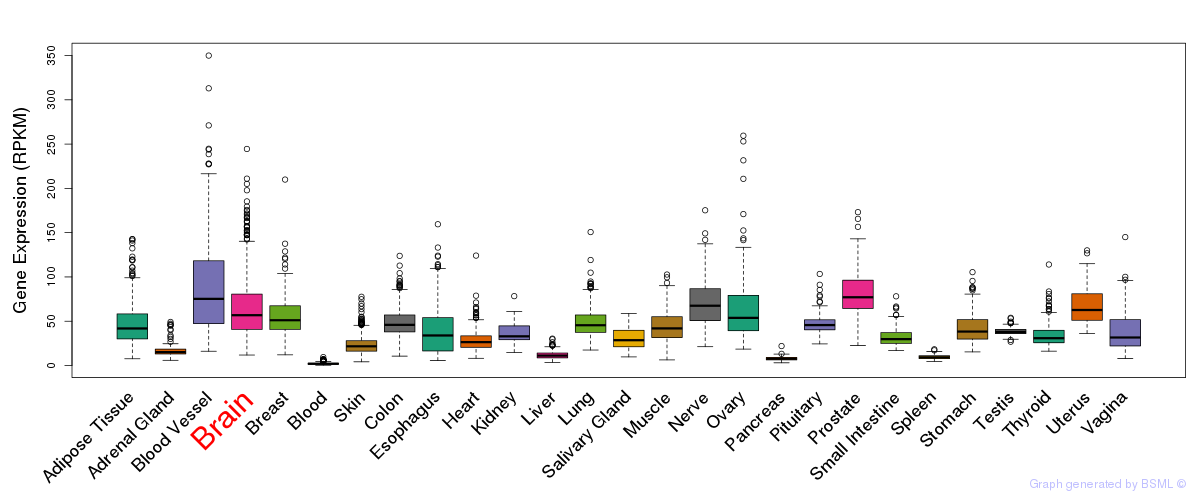
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCN8A | 0.94 | 0.93 |
| KCNB1 | 0.93 | 0.91 |
| AP000751.3 | 0.92 | 0.92 |
| MEF2D | 0.92 | 0.92 |
| DLGAP2 | 0.91 | 0.84 |
| TLN2 | 0.91 | 0.93 |
| AAK1 | 0.91 | 0.94 |
| SORL1 | 0.91 | 0.84 |
| KCNMA1 | 0.90 | 0.80 |
| LRRC8B | 0.90 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBI | -0.46 | -0.62 |
| RAB34 | -0.45 | -0.56 |
| MYL12A | -0.44 | -0.63 |
| PECI | -0.42 | -0.50 |
| RAB13 | -0.42 | -0.53 |
| RHOC | -0.42 | -0.58 |
| EFEMP2 | -0.42 | -0.44 |
| C1orf61 | -0.42 | -0.61 |
| C21orf57 | -0.41 | -0.41 |
| C9orf46 | -0.41 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 9022669 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APLP1 | APLP | amyloid beta (A4) precursor-like protein 1 | Two-hybrid | BioGRID | 16169070 |
| ASAH1 | AC | ASAH | FLJ21558 | FLJ22079 | PHP | PHP32 | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | Two-hybrid | BioGRID | 16169070 |
| BAT3 | BAG-6 | BAG6 | D6S52E | G3 | HLA-B associated transcript 3 | Affinity Capture-MS | BioGRID | 17353931 |
| CCDC90B | MDS011 | MDS025 | MGC104239 | coiled-coil domain containing 90B | Two-hybrid | BioGRID | 16169070 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| COPB2 | beta'-COP | coatomer protein complex, subunit beta 2 (beta prime) | Affinity Capture-MS | BioGRID | 17353931 |
| CORO2B | CLIPINC | KIAA0925 | coronin, actin binding protein, 2B | Two-hybrid | BioGRID | 16169070 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EPPK1 | EPIPL | EPIPL1 | epiplakin 1 | Affinity Capture-MS | BioGRID | 17353931 |
| GCN1L1 | GCN1 | GCN1L | KIAA0219 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| GSTK1 | GST13 | glutathione S-transferase kappa 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HEATR2 | FLJ20397 | FLJ25564 | FLJ31671 | FLJ39381 | HEAT repeat containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPM | DKFZp547H118 | HNRNPM4 | HNRPM | HNRPM4 | HTGR1 | NAGR1 | heterogeneous nuclear ribonucleoprotein M | Affinity Capture-MS | BioGRID | 17353931 |
| IARS | FLJ20736 | IARS1 | ILRS | PRO0785 | isoleucyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| KIAA0090 | RP1-43E13.1 | KIAA0090 | Affinity Capture-MS | BioGRID | 17353931 |
| MAP3K12 | DLK | MUK | ZPK | ZPKP1 | mitogen-activated protein kinase kinase kinase 12 | Two-hybrid | BioGRID | 16169070 |
| MTR | FLJ33168 | FLJ43216 | FLJ45386 | MS | 5-methyltetrahydrofolate-homocysteine methyltransferase | Affinity Capture-MS | BioGRID | 17353931 |
| NDUFA9 | MGC111043 | NDUFS2L | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | Affinity Capture-MS | BioGRID | 17353931 |
| NRBP1 | BCON3 | FLJ27109 | FLJ35541 | MADM | MUDPNP | NRBP | nuclear receptor binding protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| NUP93 | KIAA0095 | MGC21106 | nucleoporin 93kDa | Affinity Capture-MS | BioGRID | 17353931 |
| RGS1 | 1R20 | BL34 | IER1 | IR20 | regulator of G-protein signaling 1 | Two-hybrid | BioGRID | 16169070 |
| RPA3 | REPA3 | replication protein A3, 14kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | Two-hybrid | BioGRID | 16169070 |
| SFXN1 | FLJ12876 | sideroflexin 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SLC25A22 | FLJ13044 | GC1 | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | Affinity Capture-MS | BioGRID | 17353931 |
| SRPRB | APMCF1 | signal recognition particle receptor, B subunit | Affinity Capture-MS | BioGRID | 17353931 |
| TSC22D1 | DKFZp686O19206 | MGC17597 | RP11-269C23.2 | TGFB1I4 | TSC22 | TSC22 domain family, member 1 | Two-hybrid | BioGRID | 16169070 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | - | HPRD | 10488076 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 65 DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE DN | 51 | 38 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| WANG IMMORTALIZED BY HOXA9 AND MEIS1 DN | 24 | 15 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP | 86 | 55 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C6 | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 1 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART VENTRICLE DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO RADIATION THERAPY | 32 | 20 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C DN | 59 | 39 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G24 | 28 | 19 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 530 | 536 | m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 296 | 302 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-134 | 81 | 87 | m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-200bc/429 | 147 | 154 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 89 | 95 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-205 | 623 | 629 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-381 | 642 | 648 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-542-3p | 502 | 508 | m8 | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.