Gene Page: ASAP2
Summary ?
| GeneID | 8853 |
| Symbol | ASAP2 |
| Synonyms | AMAP2|CENTB3|DDEF2|PAG3|PAP|Pap-alpha|SHAG1 |
| Description | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| Reference | MIM:603817|HGNC:HGNC:2721|Ensembl:ENSG00000151693|HPRD:04820|Vega:OTTHUMG00000117485 |
| Gene type | protein-coding |
| Map location | 2p25|2p24 |
| Pascal p-value | 0.048 |
| Sherlock p-value | 0.576 |
| TADA p-value | 0.019 |
| Fetal beta | 0.579 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASAP2 | chr2 | 9517076 | G | A | NM_001135191 NM_003887 | p.596V>M p.596V>M | missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03424478 | 2 | 9346387 | ASAP2 | 3.85E-8 | -0.015 | 1.1E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9368942 | chr6 | 36566512 | ASAP2 | 8853 | 0.12 | trans | ||
| rs9366911 | chr6 | 36589339 | ASAP2 | 8853 | 0.07 | trans | ||
| rs7972594 | chr12 | 11724913 | ASAP2 | 8853 | 0.2 | trans | ||
| rs10491979 | chr12 | 11727969 | ASAP2 | 8853 | 0.2 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
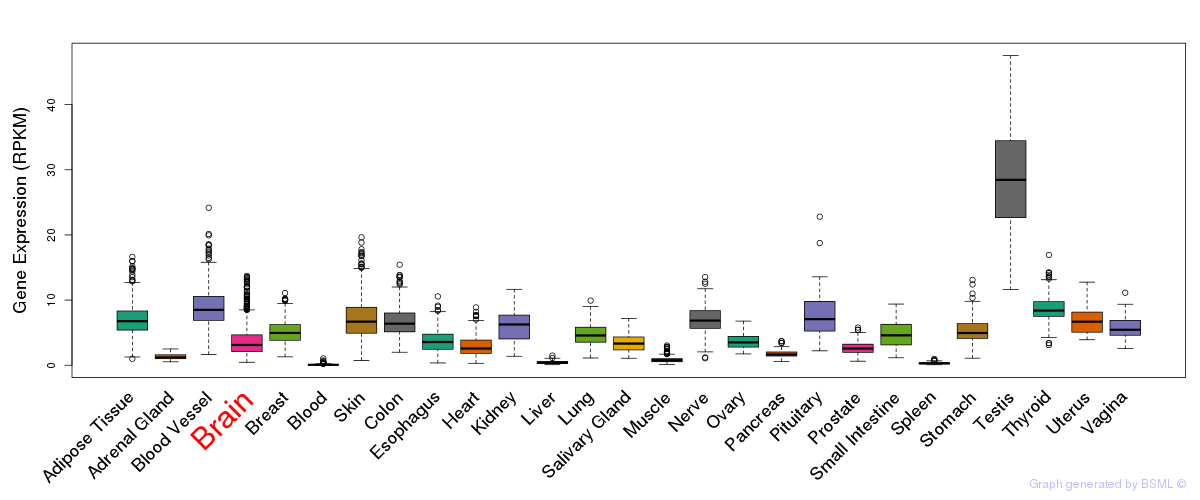
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZEB1 | 0.77 | 0.75 |
| SPAG9 | 0.76 | 0.73 |
| BMS1 | 0.75 | 0.73 |
| HDLBP | 0.75 | 0.73 |
| ZNF664 | 0.75 | 0.69 |
| LRBA | 0.75 | 0.74 |
| B3GALNT2 | 0.75 | 0.74 |
| NBR1 | 0.74 | 0.73 |
| CDYL | 0.74 | 0.71 |
| PDCD4 | 0.74 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.57 | -0.47 |
| C1orf54 | -0.54 | -0.48 |
| VAMP5 | -0.51 | -0.45 |
| IFI27 | -0.50 | -0.45 |
| AF347015.31 | -0.50 | -0.45 |
| IL32 | -0.50 | -0.46 |
| MYL3 | -0.49 | -0.45 |
| MT-CO2 | -0.49 | -0.43 |
| SYCP3 | -0.49 | -0.38 |
| ENHO | -0.48 | -0.49 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARF1 | - | ADP-ribosylation factor 1 | - | HPRD | 10022920 |10749932 |
| ARF5 | - | ADP-ribosylation factor 5 | - | HPRD | 10022920 |10749932 |
| ARF6 | DKFZp564M0264 | ADP-ribosylation factor 6 | - | HPRD | 10022920 |10749932 |
| ITSN1 | ITSN | MGC134948 | MGC134949 | SH3D1A | SH3P17 | intersectin 1 (SH3 domain protein) | DDEF2 (AMAP2) interacts with an unspecified isoform of ITSN1 (Intersectin1). | BIND | 15719014 |
| PACSIN3 | SDPIII | protein kinase C and casein kinase substrate in neurons 3 | DDEF2 (AMAP2) interacts with PACSIN3. | BIND | 15719014 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10022920 |
| PXN | FLJ16691 | paxillin | in vitro Reconstituted Complex | BioGRID | 10749932 |12149250 |
| PXN | FLJ16691 | paxillin | - | HPRD | 10749932|12149250 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | - | HPRD | 10022920 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | DDEF2 (AMAP2) interacts with SH3KBP1 (CIN85). | BIND | 15719014 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 10022920 |
| TBRG4 | CPR2 | FASTKD4 | KIAA0948 | transforming growth factor beta regulator 4 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARAP PATHWAY | 18 | 13 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 6HR | 26 | 15 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |