Gene Page: WASF1
Summary ?
| GeneID | 8936 |
| Symbol | WASF1 |
| Synonyms | SCAR1|WAVE|WAVE1 |
| Description | WAS protein family member 1 |
| Reference | MIM:605035|HGNC:HGNC:12732|Ensembl:ENSG00000112290|HPRD:05434|Vega:OTTHUMG00000015357 |
| Gene type | protein-coding |
| Map location | 6q21 |
| Pascal p-value | 0.096 |
| Sherlock p-value | 0.804 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.11:CC_BA10_disease_P=0.0291:HBB_BA9_fold_change=-1.15:HBB_BA9_disease_P=0.0091 |
| Fetal beta | 0.117 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-1983537 | 0 | WASF1 | 8936 | 0.13 | trans |
Section II. Transcriptome annotation
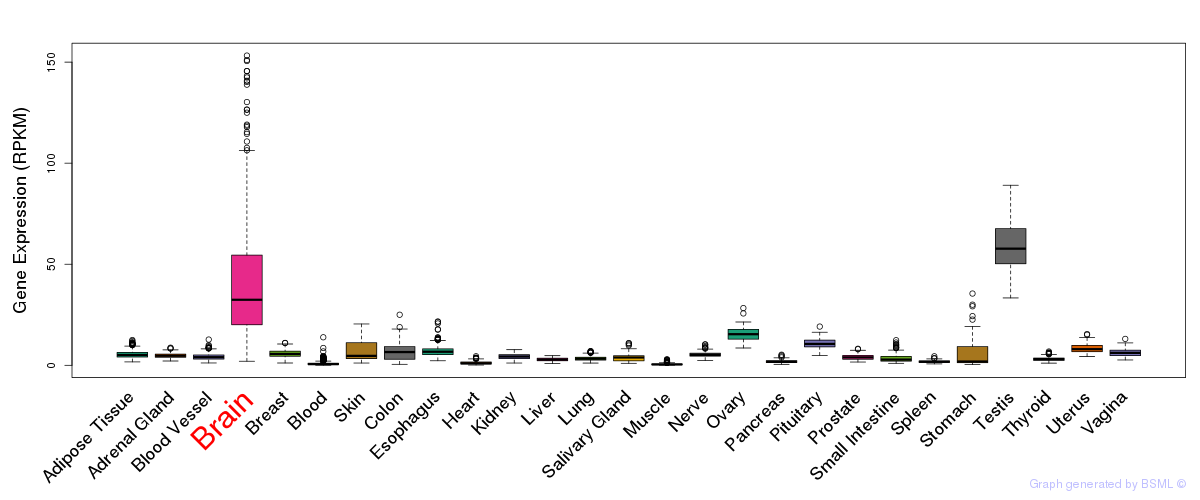
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYK | 0.73 | 0.65 |
| FCGR3A | 0.73 | 0.66 |
| VSIG4 | 0.73 | 0.72 |
| NCKAP1L | 0.73 | 0.71 |
| CD68 | 0.72 | 0.73 |
| FCGR1A | 0.72 | 0.57 |
| HCK | 0.72 | 0.65 |
| SIGLEC10 | 0.71 | 0.65 |
| ITGB2 | 0.70 | 0.68 |
| FPR1 | 0.70 | 0.56 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.27 | -0.34 |
| AF347015.26 | -0.26 | -0.34 |
| AF347015.18 | -0.26 | -0.36 |
| AF347015.8 | -0.26 | -0.33 |
| AF347015.2 | -0.25 | -0.35 |
| AF347015.33 | -0.25 | -0.31 |
| AF347015.27 | -0.25 | -0.32 |
| AC098691.2 | -0.24 | -0.32 |
| MT-CYB | -0.24 | -0.31 |
| AF347015.31 | -0.23 | -0.31 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | WAVE-1 interacts with Abl. | BIND | 10970852 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 10970852 |
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | WAVE-1 interacts with G-actin. This interaction was modeled on a demonstrated interaction between human WAVE-1 and rat G-actin. | BIND | 10970852 |
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | Affinity Capture-Western | BioGRID | 9843499 |
| BAIAP2 | BAP2 | IRSP53 | BAI1-associated protein 2 | Far Western Reconstituted Complex Two-hybrid | BioGRID | 11130076 |
| C3orf10 | HSPC300 | MDS027 | hHBrk1 | chromosome 3 open reading frame 10 | - | HPRD | 12181570 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD | 12181570 |
| DNMBP | KIAA1010 | TUBA | dynamin binding protein | - | HPRD,BioGRID | 14506234 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD | 10532312 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 9307968 |
| PFN1 | - | profilin 1 | - | HPRD,BioGRID | 9843499 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | WAVE-1 interacts with PKA. | BIND | 10970852 |
| PSTPIP1 | CD2BP1 | CD2BP1L | CD2BP1S | H-PIP | PAPAS | PSTPIP | proline-serine-threonine phosphatase interacting protein 1 | - | HPRD | 9804817 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD | 9843499 |
| SRGAP3 | ARHGAP14 | KIAA0411 | MEGAP | SRGAP2 | WRP | SLIT-ROBO Rho GTPase activating protein 3 | - | HPRD,BioGRID | 12447388 |
| SRGAP3 | ARHGAP14 | KIAA0411 | MEGAP | SRGAP2 | WRP | SLIT-ROBO Rho GTPase activating protein 3 | WAVE1 interacts with WRP. This interaction was modeled on a demonstrated interaction between WAVE-1 from rat and an unspecified sources and WRP from rat, human and an unspecified sources. | BIND | 12447388 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SALMONELLA PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAC1 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACTINY PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| SIG CHEMOTAXIS | 45 | 37 | All SZGR 2.0 genes in this pathway |
| SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | 35 | 25 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA DN | 16 | 8 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |