Gene Page: CCNT1
Summary ?
| GeneID | 904 |
| Symbol | CCNT1 |
| Synonyms | CCNT|CYCT1|HIVE1 |
| Description | cyclin T1 |
| Reference | MIM:143055|HGNC:HGNC:1599|Ensembl:ENSG00000129315|HPRD:03938|Vega:OTTHUMG00000170393 |
| Gene type | protein-coding |
| Map location | 12q13.11 |
| Pascal p-value | 0.386 |
| Sherlock p-value | 0.562 |
| Fetal beta | 0.954 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.606 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22103585 | 12 | 49110427 | CCNT1 | -0.032 | 0.46 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9637082 | chr21 | 21213235 | CCNT1 | 904 | 0.14 | trans |
Section II. Transcriptome annotation
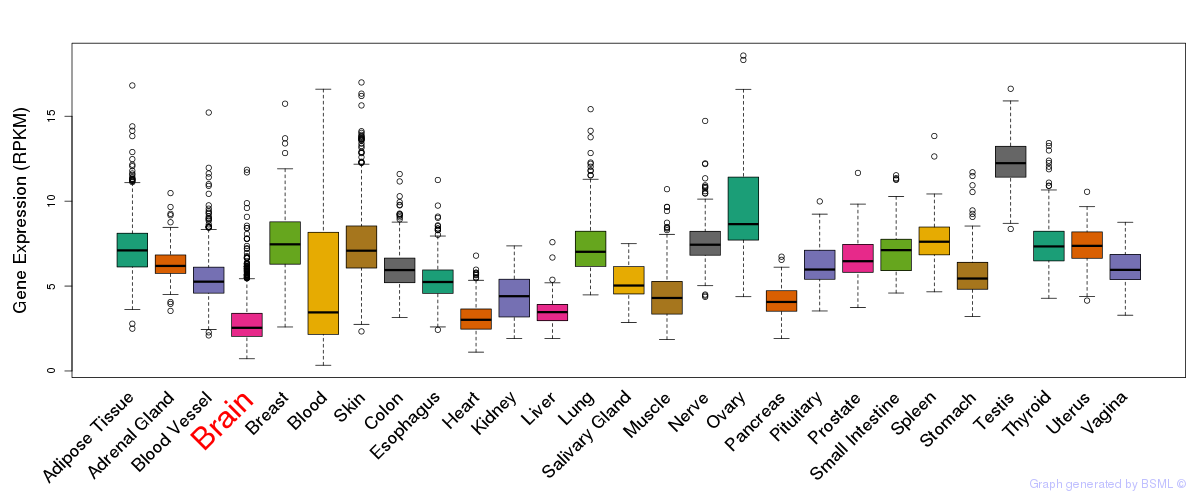
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IDA | 16109376 | |
| GO:0005515 | protein binding | IPI | 12944466 | |
| GO:0017069 | snRNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000079 | regulation of cyclin-dependent protein kinase activity | TAS | 9499409 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IPI | 16109376 | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 9499409 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0051301 | cell division | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 12944466 | |
| GO:0005654 | nucleoplasm | EXP | 7853496 |12676794 | |
| GO:0005730 | nucleolus | IDA | 16109376 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AFF4 | AF5Q31 | MCEF | MGC75036 | AF4/FMR2 family, member 4 | - | HPRD,BioGRID | 12065898 |
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 12917420 |
| CCNK | CPR4 | MGC9113 | cyclin K | in vitro in vivo | BioGRID | 10574912 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | - | HPRD | 9872325 |10465067 |10617616 |11145967 |11689688 |11713532 |11713533 |12115727 |12894230 |14627702 |15528190 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | CyclinT1 interacts with Cdk9. | BIND | 9499409 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | CycT1 interacts with CDK9. This interaction was modeled on a demonstrated interaction between CycT1 from an unspecified species and human CDK9. | BIND | 15546612 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | Affinity Capture-Western in vitro in vivo Phenotypic Suppression Reconstituted Complex Two-hybrid | BioGRID | 9499409 |10656684 |10958691 |11689688 |12588988 |12832472 |12944466 |15107825 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | Cyclin T1 interacts with CDK9. | BIND | 12588988 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | - | HPRD | 9499409 |10958691 |
| GRIPAP1 | DKFZp434P0630 | GRASP-1 | KIAA1167 | MGC126593 | MGC126595 | GRIP1 associated protein 1 | in vitro in vivo | BioGRID | 11704662 |
| GRN | GEP | GP88 | PCDGF | PEPI | PGRN | granulin | - | HPRD,BioGRID | 12588988 |
| GRN | GEP | GP88 | PCDGF | PEPI | PGRN | granulin | Cyclin T1 interacts with granulin. | BIND | 12588988 |
| HEXIM1 | CLP1 | EDG1 | FLJ13562 | HIS1 | MAQ1 | hexamethylene bis-acetamide inducible 1 | - | HPRD,BioGRID | 12832472 |
| HTATSF1 | TAT-SF1 | dJ196E23.2 | HIV-1 Tat specific factor 1 | - | HPRD,BioGRID | 10913173 |11780068 |
| MDFIC | HIC | MyoD family inhibitor domain containing | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12944466 |
| MYBL2 | B-MYB | BMYB | MGC15600 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | Phenotypic Suppression | BioGRID | 10656684 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 12944920 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | Cyclin T1 interacts with GRIP1. | BIND | 11704662 |
| NUFIP1 | NUFIP | bA540M5.1 | nuclear fragile X mental retardation protein interacting protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15107825 |
| NUFIP1 | NUFIP | bA540M5.1 | nuclear fragile X mental retardation protein interacting protein 1 | CyclinT1 interacts with NUFIP. | BIND | 15107825 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 12727882 |
| PURA | PUR-ALPHA | PUR1 | PURALPHA | purine-rich element binding protein A | - | HPRD,BioGRID | 11730934 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | - | HPRD | 11689688 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME ELONGATION ARREST AND RECOVERY | 32 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| NOUZOVA TRETINOIN AND H4 ACETYLATION | 143 | 85 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS DN | 108 | 64 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT1 TARGETS | 50 | 26 | All SZGR 2.0 genes in this pathway |
| LE NEURONAL DIFFERENTIATION DN | 19 | 16 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |