Gene Page: MTMR6
Summary ?
| GeneID | 9107 |
| Symbol | MTMR6 |
| Synonyms | - |
| Description | myotubularin related protein 6 |
| Reference | MIM:603561|HGNC:HGNC:7453|Ensembl:ENSG00000139505|HPRD:11946|Vega:OTTHUMG00000016606 |
| Gene type | protein-coding |
| Map location | 13q12 |
| Pascal p-value | 0.504 |
| Sherlock p-value | 0.892 |
| Fetal beta | -0.123 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24867566 | 13 | 25861182 | MTMR6 | 6.43E-8 | -0.011 | 1.59E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4491879 | chr3 | 189373908 | MTMR6 | 9107 | 0.14 | trans |
Section II. Transcriptome annotation
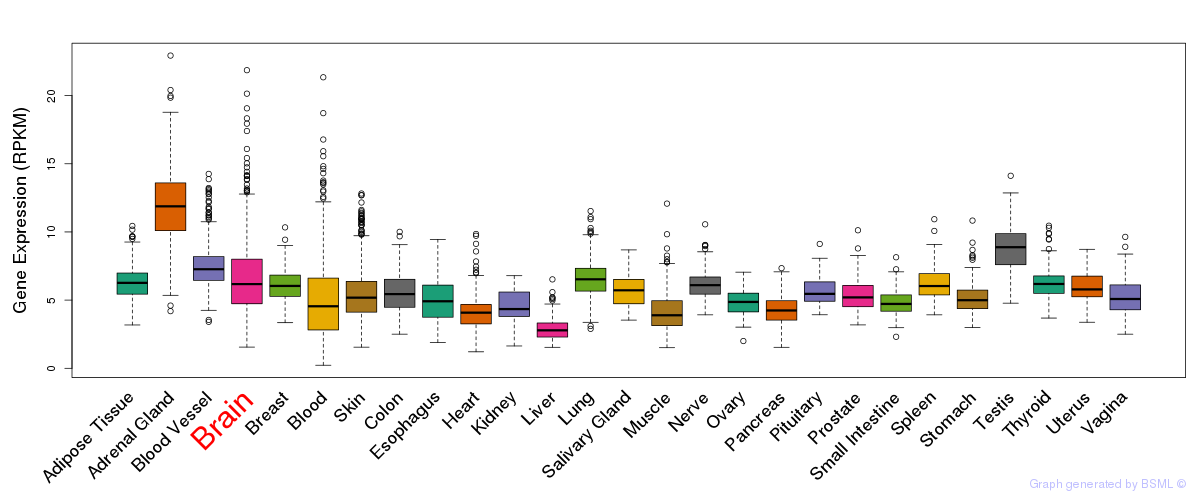
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC087645.2 | 0.93 | 0.54 |
| CDCA3 | 0.92 | 0.54 |
| BIRC5 | 0.92 | 0.53 |
| UBE2C | 0.92 | 0.48 |
| AURKB | 0.92 | 0.46 |
| FAM64A | 0.91 | 0.53 |
| CDC45L | 0.91 | 0.47 |
| TACC3 | 0.91 | 0.59 |
| ZWINT | 0.91 | 0.68 |
| CCNB2 | 0.91 | 0.50 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.36 | -0.46 |
| AF347015.33 | -0.35 | -0.44 |
| AF347015.15 | -0.35 | -0.48 |
| MT-CO2 | -0.35 | -0.43 |
| MT-CYB | -0.35 | -0.45 |
| AF347015.8 | -0.35 | -0.46 |
| TINAGL1 | -0.34 | -0.38 |
| AF347015.26 | -0.34 | -0.48 |
| AF347015.31 | -0.34 | -0.39 |
| AF347015.2 | -0.33 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004725 | protein tyrosine phosphatase activity | NAS | 9736772 | |
| GO:0004722 | protein serine/threonine phosphatase activity | NAS | 9736772 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006470 | protein amino acid dephosphorylation | NAS | 9736772 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | 9736772 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005635 | nuclear envelope | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 11256614 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FRUCTOSE AND MANNOSE METABOLISM | 34 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG RIBOFLAVIN METABOLISM | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME PI METABOLISM | 48 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D | 68 | 44 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 484 | 490 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-183 | 458 | 464 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-496 | 1909 | 1916 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.