Gene Page: BCL7B
Summary ?
| GeneID | 9275 |
| Symbol | BCL7B |
| Synonyms | - |
| Description | B-cell CLL/lymphoma 7B |
| Reference | MIM:605846|HGNC:HGNC:1005|Ensembl:ENSG00000106635|HPRD:05786|Vega:OTTHUMG00000023412 |
| Gene type | protein-coding |
| Map location | 7q11.23 |
| Pascal p-value | 0.14 |
| Sherlock p-value | 0.625 |
| Fetal beta | -1.179 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10891578 | chr11 | 113522710 | BCL7B | 9275 | 0.11 | trans | ||
| rs1715395 | chr20 | 4931591 | BCL7B | 9275 | 0.08 | trans |
Section II. Transcriptome annotation
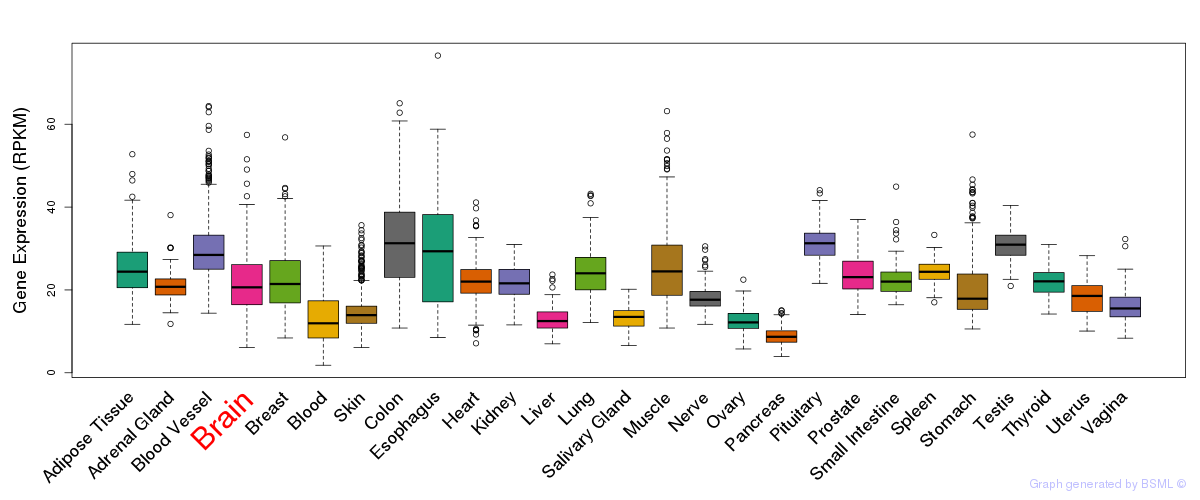
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC8C | 0.78 | 0.81 |
| SPRED1 | 0.77 | 0.77 |
| LEPR | 0.76 | 0.76 |
| CD164 | 0.75 | 0.80 |
| FRMD6 | 0.74 | 0.76 |
| TEK | 0.74 | 0.80 |
| SLC30A1 | 0.73 | 0.79 |
| RASSF8 | 0.73 | 0.80 |
| PCGF5 | 0.72 | 0.75 |
| DPP8 | 0.71 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNHG12 | -0.45 | -0.54 |
| AL022328.1 | -0.43 | -0.43 |
| POLR2J | -0.41 | -0.50 |
| BCL7C | -0.41 | -0.51 |
| AC132872.1 | -0.40 | -0.48 |
| RPS21 | -0.40 | -0.53 |
| NME4 | -0.39 | -0.49 |
| RPL28 | -0.39 | -0.54 |
| AP003068.3 | -0.39 | -0.42 |
| RP9P | -0.39 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR UP | 47 | 39 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES DN | 30 | 19 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |