Gene Page: ACVRL1
Summary ?
| GeneID | 94 |
| Symbol | ACVRL1 |
| Synonyms | ACVRLK1|ALK-1|ALK1|HHT|HHT2|ORW2|SKR3|TSR-I |
| Description | activin A receptor like type 1 |
| Reference | MIM:601284|HGNC:HGNC:175|Ensembl:ENSG00000139567|HPRD:03181|Vega:OTTHUMG00000169507 |
| Gene type | protein-coding |
| Map location | 12q13.13 |
| Pascal p-value | 0.502 |
| Fetal beta | -0.214 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12193731 | 12 | 52300911 | ACVRL1 | 4.312E-4 | -0.642 | 0.045 | DMG:Wockner_2014 |
| cg09043511 | 12 | 52305945 | ACVRL1 | 5.952E-4 | 0.321 | 0.05 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2190759 | chr17 | 55794698 | ACVRL1 | 94 | 0.19 | trans |
Section II. Transcriptome annotation
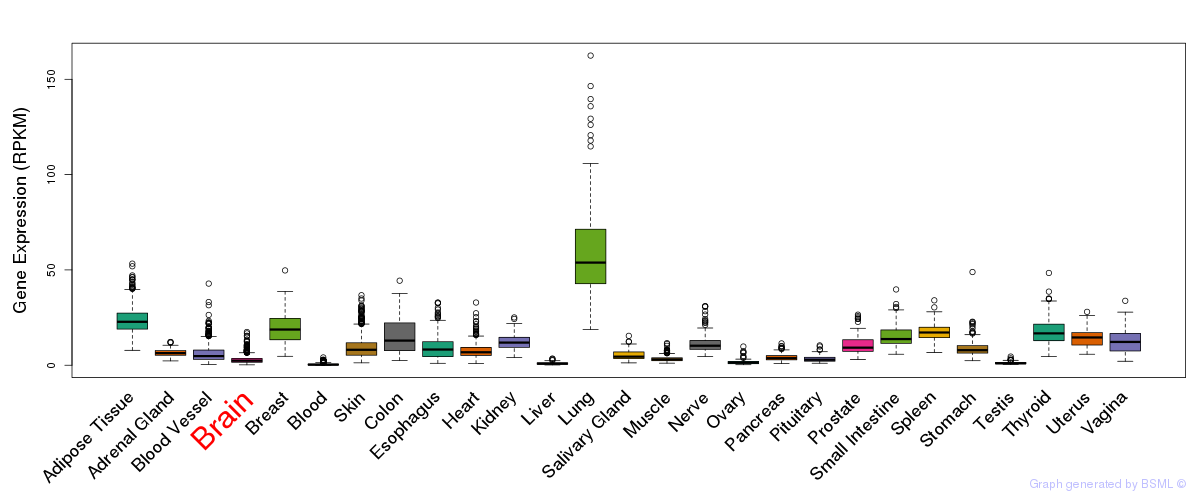
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IFT20 | 0.71 | 0.77 |
| FKBP3 | 0.70 | 0.74 |
| SNRPA1 | 0.70 | 0.77 |
| AC022098.3 | 0.70 | 0.58 |
| FAM50A | 0.70 | 0.72 |
| PDCD5 | 0.69 | 0.72 |
| GRK4 | 0.69 | 0.68 |
| GCHFR | 0.69 | 0.69 |
| NDUFS4 | 0.68 | 0.72 |
| ANAPC4 | 0.68 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.52 | -0.58 |
| MT-CYB | -0.49 | -0.53 |
| AF347015.15 | -0.49 | -0.53 |
| AF347015.31 | -0.49 | -0.51 |
| AF347015.27 | -0.48 | -0.53 |
| AF347015.8 | -0.48 | -0.51 |
| MT-CO2 | -0.48 | -0.50 |
| AF347015.33 | -0.47 | -0.50 |
| AF347015.26 | -0.47 | -0.54 |
| PRELP | -0.46 | -0.50 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL RUNX1 | 9 | 6 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN | 48 | 33 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER BY MUTATION RATE | 20 | 18 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |