Gene Page: CHD1L
Summary ?
| GeneID | 9557 |
| Symbol | CHD1L |
| Synonyms | ALC1|CHDL |
| Description | chromodomain helicase DNA binding protein 1-like |
| Reference | MIM:613039|HGNC:HGNC:1916|Ensembl:ENSG00000131778|HPRD:09880|Vega:OTTHUMG00000150271 |
| Gene type | protein-coding |
| Map location | 1q12 |
| Sherlock p-value | 0.196 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24197750 | 1 | 146714116 | CHD1L | 8.9E-8 | -0.007 | 2.04E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs74125727 | 1 | 147017714 | CHD1L | ENSG00000131778.13 | 3.06419E-6 | 0.03 | 293436 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
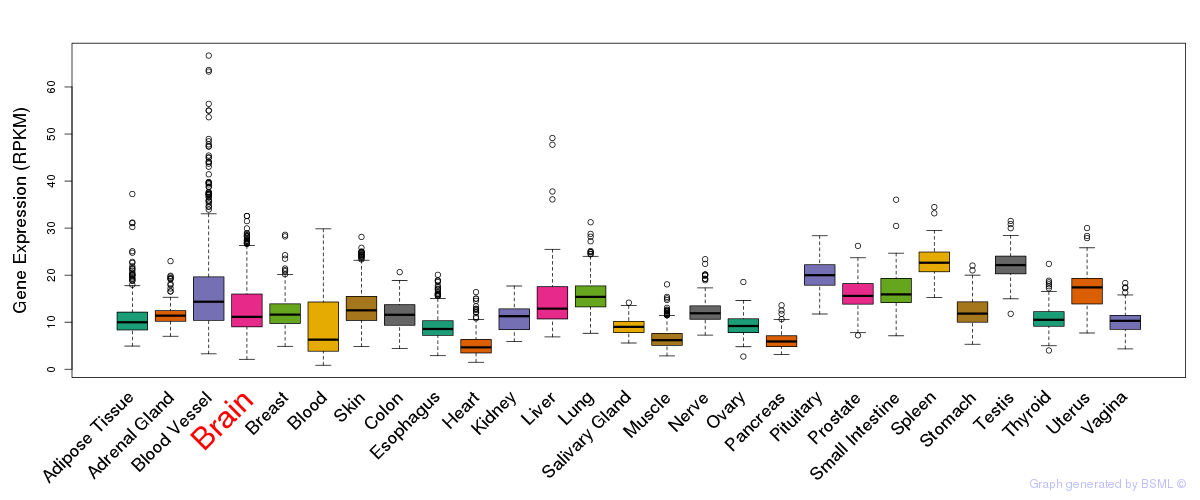
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BRSK2 | 0.92 | 0.92 |
| MBD1 | 0.92 | 0.92 |
| HGS | 0.91 | 0.91 |
| ANAPC2 | 0.91 | 0.90 |
| ARFGAP1 | 0.91 | 0.91 |
| NCLN | 0.91 | 0.92 |
| RNF112 | 0.91 | 0.91 |
| DHX30 | 0.91 | 0.89 |
| IGHMBP2 | 0.90 | 0.90 |
| HIRA | 0.90 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.74 |
| AF347015.21 | -0.78 | -0.79 |
| MT-CO2 | -0.77 | -0.73 |
| AF347015.33 | -0.76 | -0.71 |
| AF347015.8 | -0.75 | -0.73 |
| AF347015.27 | -0.75 | -0.74 |
| MT-CYB | -0.74 | -0.70 |
| AF347015.2 | -0.72 | -0.67 |
| HIGD1B | -0.71 | -0.70 |
| AF347015.15 | -0.71 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008026 | ATP-dependent helicase activity | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| VISALA RESPONSE TO HEAT SHOCK AND AGING DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |