Gene Page: CD40LG
Summary ?
| GeneID | 959 |
| Symbol | CD40LG |
| Synonyms | CD154|CD40L|HIGM1|IGM|IMD3|T-BAM|TNFSF5|TRAP|gp39|hCD40L |
| Description | CD40 ligand |
| Reference | MIM:300386|HGNC:HGNC:11935|Ensembl:ENSG00000102245|HPRD:02311|Vega:OTTHUMG00000022512 |
| Gene type | protein-coding |
| Map location | Xq26 |
| Fetal beta | -0.488 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02992767 | X | 135729973 | CD40LG | 0.003 | -5.026 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
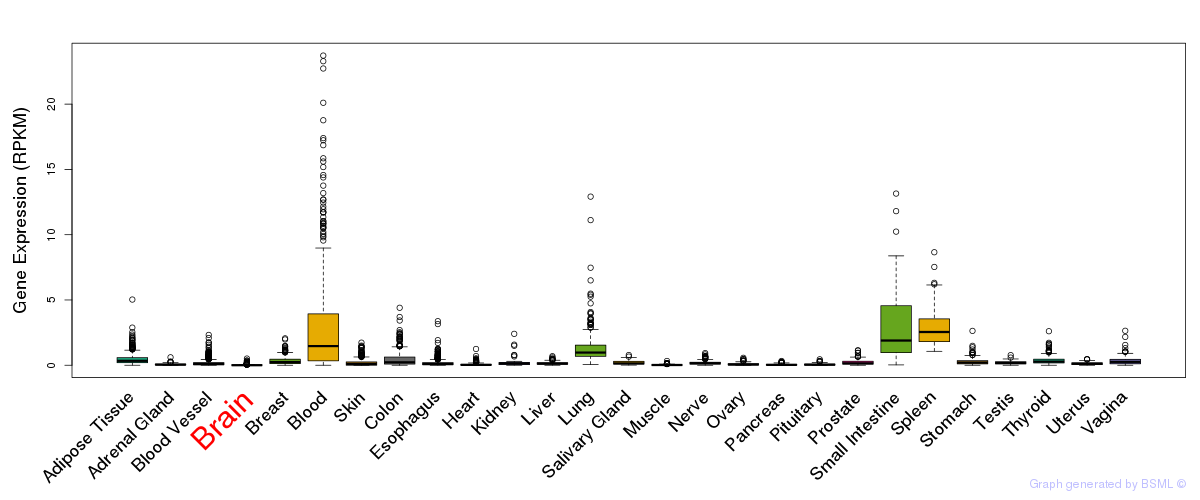
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION | 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE | 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG PRIMARY IMMUNODEFICIENCY | 35 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ASBCELL PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CD40 PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TH1TH2 PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER DN | 59 | 45 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS RESISTANT UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA DN | 45 | 28 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS AND CYCLIC RGD | 21 | 15 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |