Gene Page: CD81
Summary ?
| GeneID | 975 |
| Symbol | CD81 |
| Synonyms | CVID6|S5.7|TAPA1|TSPAN28 |
| Description | CD81 molecule |
| Reference | MIM:186845|HGNC:HGNC:1701|Ensembl:ENSG00000110651|HPRD:08924|Vega:OTTHUMG00000009892 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.298 |
| Sherlock p-value | 0.769 |
| Fetal beta | -0.622 |
| DMG | 2 (# studies) |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 6 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 6 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20321801 | 11 | 2404384 | CD81 | 1.46E-5 | -0.436 | 0.015 | DMG:Wockner_2014 |
| cg26382697 | 11 | 2406712 | CD81 | 2.391E-4 | 0.46 | 0.037 | DMG:Wockner_2014 |
| cg16465939 | 11 | 2554410 | CD81 | 2.19E-5 | 5.899 | DMG:vanEijk_2014 | |
| cg17627559 | 11 | 2323896 | CD81 | 8.99E-6 | 5.499 | DMG:vanEijk_2014 | |
| cg05656180 | 11 | 2890258 | CD81 | 0.001 | 4.823 | DMG:vanEijk_2014 | |
| cg12949760 | 11 | 2542862 | CD81 | 1.54E-5 | 4.647 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
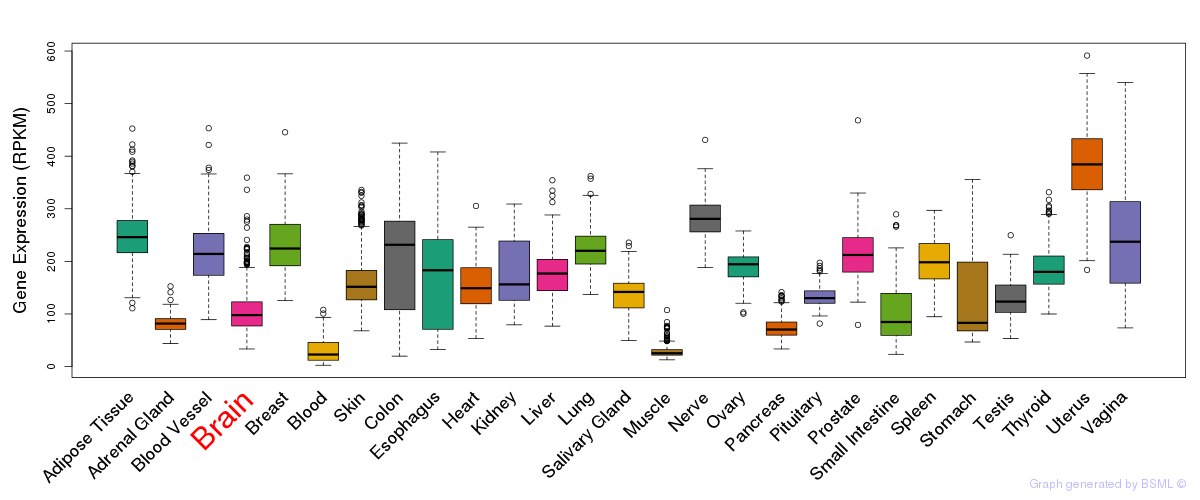
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD19 | B4 | MGC12802 | CD19 molecule | Affinity Capture-Western | BioGRID | 1383329 |7636191 |9804823 |
| CD4 | CD4mut | CD4 molecule | Affinity Capture-Western | BioGRID | 7636191 |
| CD46 | MCP | MGC26544 | MIC10 | TLX | TRA2.10 | CD46 molecule, complement regulatory protein | Affinity Capture-Western | BioGRID | 10741407 |
| CD63 | LAMP-3 | ME491 | MLA1 | OMA81H | TSPAN30 | CD63 molecule | - | HPRD | 12036870 |
| CD63 | LAMP-3 | ME491 | MLA1 | OMA81H | TSPAN30 | CD63 molecule | in vivo | BioGRID | 9006891 |
| CD81 | S5.7 | TAPA1 | TSPAN28 | CD81 molecule | - | HPRD,BioGRID | 12437138 |
| CD9 | 5H9 | BA2 | BTCC-1 | DRAP-27 | GIG2 | MIC3 | MRP-1 | P24 | TSPAN29 | CD9 molecule | - | HPRD | 12036870 |
| CD9 | 5H9 | BA2 | BTCC-1 | DRAP-27 | GIG2 | MIC3 | MRP-1 | P24 | TSPAN29 | CD9 molecule | Affinity Capture-Western Co-fractionation | BioGRID | 8630057 |9804823 |
| CR2 | C3DR | CD21 | SLEB9 | complement component (3d/Epstein Barr virus) receptor 2 | Affinity Capture-Western | BioGRID | 1383329 |
| DLG5 | KIAA0583 | LP-DLG | P-DLG5 | PDLG | discs, large homolog 5 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| IFITM1 | 9-27 | CD225 | IFI17 | LEU13 | interferon induced transmembrane protein 1 (9-27) | Affinity Capture-Western | BioGRID | 2398277 |
| IGSF8 | CD316 | CD81P3 | EWI2 | PGRL | immunoglobulin superfamily, member 8 | Affinity Capture-Western | BioGRID | 11673522 |
| INO80B | HMGA1L4 | HMGIYL4 | IES2 | PAP-1BP | PAPA-1 | ZNHIT4 | hIes2 | INO80 complex subunit B | Two-hybrid | BioGRID | 16169070 |
| ITGA4 | CD49D | IA4 | MGC90518 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | - | HPRD,BioGRID | 10229664 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD,BioGRID | 10229664 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 12036870 |
| PTGFRN | CD315 | CD9P-1 | EWI-F | FLJ11001 | FPRP | KIAA1436 | SMAP-6 | prostaglandin F2 receptor negative regulator | Affinity Capture-Western in vitro in vivo | BioGRID | 11087758 |11278880 |
| TSPAN4 | NAG-2 | NAG2 | TETRASPAN | TM4SF7 | TSPAN-4 | tetraspanin 4 | Affinity Capture-Western | BioGRID | 9360996 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 | 71 | 42 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| LEIN OLIGODENDROCYTE MARKERS | 74 | 53 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| HANN RESISTANCE TO BCL2 INHIBITOR UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| MATZUK FERTILIZATION | 8 | 5 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE UP | 79 | 40 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 9 | 35 | 26 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| GERHOLD RESPONSE TO TZD DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION TOP20 DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |