Gene Page: ACYP2
Summary ?
| GeneID | 98 |
| Symbol | ACYP2 |
| Synonyms | ACYM|ACYP |
| Description | acylphosphatase 2 |
| Reference | MIM:102595|HGNC:HGNC:180|Ensembl:ENSG00000170634|HPRD:00028|Vega:OTTHUMG00000129287 |
| Gene type | protein-coding |
| Map location | 2p16.2 |
| Pascal p-value | 0.029 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
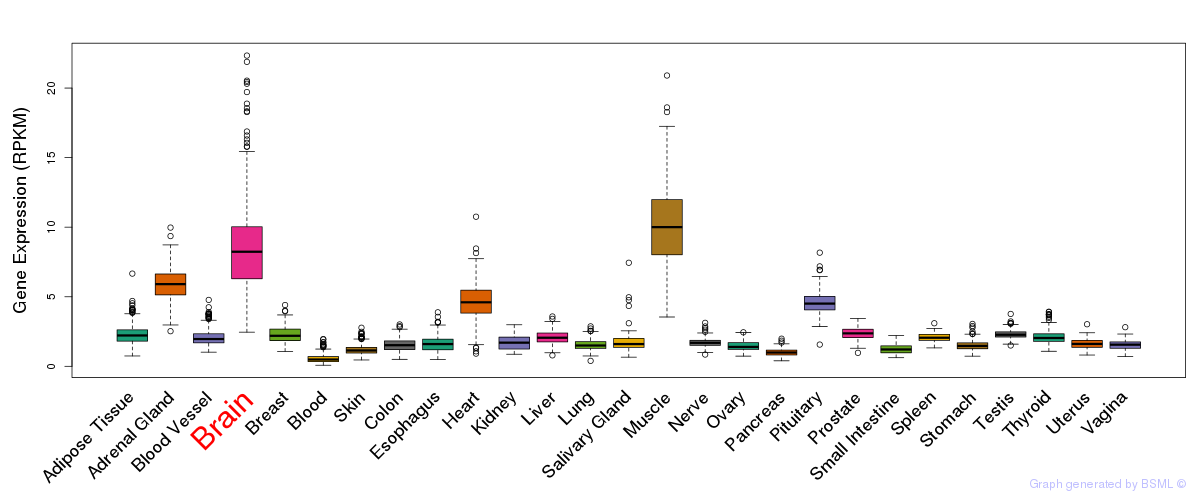
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PILRB | 0.78 | 0.80 |
| CPT1B | 0.76 | 0.80 |
| CTB-161A2.2 | 0.74 | 0.77 |
| DNASE1L2 | 0.74 | 0.73 |
| TNFRSF25 | 0.74 | 0.79 |
| RP3-402G11.1 | 0.73 | 0.72 |
| MAMDC4 | 0.73 | 0.78 |
| GNB3 | 0.73 | 0.75 |
| PRICKLE4 | 0.73 | 0.74 |
| JMJD7-PLA2G4B | 0.73 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ITM2A | -0.35 | -0.37 |
| GNG11 | -0.34 | -0.38 |
| AF347015.31 | -0.34 | -0.29 |
| TM4SF18 | -0.34 | -0.33 |
| IGFBP7 | -0.33 | -0.33 |
| AF347015.21 | -0.33 | -0.35 |
| LY96 | -0.33 | -0.41 |
| NMI | -0.32 | -0.34 |
| GIMAP2 | -0.32 | -0.33 |
| C18orf32 | -0.32 | -0.42 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PYRUVATE METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| NADLER HYPERGLYCEMIA AT OBESITY | 58 | 35 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 36HR | 29 | 23 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |