Gene Page: GFPT2
Summary ?
| GeneID | 9945 |
| Symbol | GFPT2 |
| Synonyms | GFAT|GFAT 2|GFAT2 |
| Description | glutamine-fructose-6-phosphate transaminase 2 |
| Reference | MIM:603865|HGNC:HGNC:4242|Ensembl:ENSG00000131459|HPRD:04842|Vega:OTTHUMG00000163442 |
| Gene type | protein-coding |
| Map location | 5q34-q35 |
| Pascal p-value | 0.65 |
| Sherlock p-value | 0.731 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23260877 | 5 | 179742954 | GFPT2 | 2.207E-4 | -0.527 | 0.036 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs76025377 | 5 | 179722398 | GFPT2 | ENSG00000131459.8 | 5.549E-7 | 0.01 | 57989 | gtex_brain_putamen_basal |
| rs6866031 | 5 | 179724471 | GFPT2 | ENSG00000131459.8 | 2.932E-7 | 0.01 | 55916 | gtex_brain_putamen_basal |
| rs7711688 | 5 | 179725224 | GFPT2 | ENSG00000131459.8 | 1.893E-7 | 0.01 | 55163 | gtex_brain_putamen_basal |
| rs7728939 | 5 | 179725264 | GFPT2 | ENSG00000131459.8 | 1.893E-7 | 0.01 | 55123 | gtex_brain_putamen_basal |
| rs11748698 | 5 | 179727677 | GFPT2 | ENSG00000131459.8 | 6.505E-7 | 0.01 | 52710 | gtex_brain_putamen_basal |
| rs7725 | 5 | 179727957 | GFPT2 | ENSG00000131459.8 | 6.754E-7 | 0.01 | 52430 | gtex_brain_putamen_basal |
| rs11740631 | 5 | 179731113 | GFPT2 | ENSG00000131459.8 | 4.935E-7 | 0.01 | 49274 | gtex_brain_putamen_basal |
| rs77017168 | 5 | 179731297 | GFPT2 | ENSG00000131459.8 | 4.935E-7 | 0.01 | 49090 | gtex_brain_putamen_basal |
| rs57931856 | 5 | 179731328 | GFPT2 | ENSG00000131459.8 | 4.935E-7 | 0.01 | 49059 | gtex_brain_putamen_basal |
| rs3763131 | 5 | 179734312 | GFPT2 | ENSG00000131459.8 | 4.934E-7 | 0.01 | 46075 | gtex_brain_putamen_basal |
| rs888924 | 5 | 179738213 | GFPT2 | ENSG00000131459.8 | 6.894E-7 | 0.01 | 42174 | gtex_brain_putamen_basal |
| rs116634077 | 5 | 179739034 | GFPT2 | ENSG00000131459.8 | 1.342E-7 | 0.01 | 41353 | gtex_brain_putamen_basal |
| rs75664354 | 5 | 179740108 | GFPT2 | ENSG00000131459.8 | 1.141E-6 | 0.01 | 40279 | gtex_brain_putamen_basal |
| rs11744073 | 5 | 179740239 | GFPT2 | ENSG00000131459.8 | 1.262E-6 | 0.01 | 40148 | gtex_brain_putamen_basal |
| rs190440456 | 5 | 179740527 | GFPT2 | ENSG00000131459.8 | 5.69E-7 | 0.01 | 39860 | gtex_brain_putamen_basal |
| rs2303007 | 5 | 179740827 | GFPT2 | ENSG00000131459.8 | 1.321E-6 | 0.01 | 39560 | gtex_brain_putamen_basal |
| rs77381392 | 5 | 179745413 | GFPT2 | ENSG00000131459.8 | 6.343E-7 | 0.01 | 34974 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
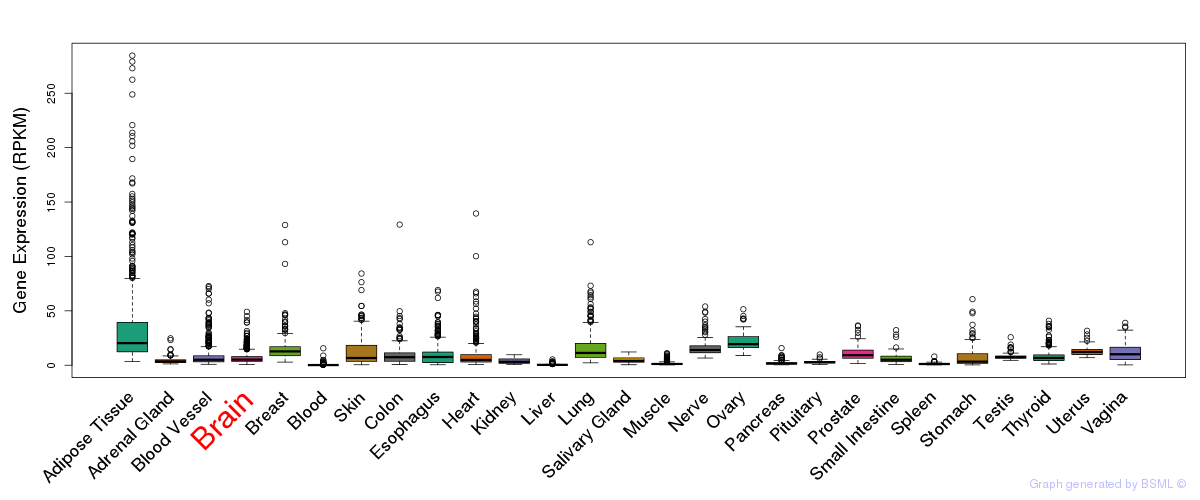
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARHGAP20 | 0.77 | 0.80 |
| JAZF1 | 0.76 | 0.78 |
| TM6SF1 | 0.75 | 0.79 |
| HIVEP2 | 0.74 | 0.77 |
| DLG2 | 0.74 | 0.80 |
| PRPS2 | 0.74 | 0.79 |
| PTCHD1 | 0.74 | 0.77 |
| PCDHA5 | 0.74 | 0.75 |
| CDH10 | 0.74 | 0.79 |
| PVRL3 | 0.74 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C11orf67 | -0.49 | -0.51 |
| AC098691.2 | -0.47 | -0.54 |
| RAB34 | -0.46 | -0.51 |
| FXYD1 | -0.46 | -0.38 |
| AP002478.3 | -0.45 | -0.47 |
| ACSF2 | -0.45 | -0.46 |
| SLC2A4RG | -0.45 | -0.45 |
| AC021016.1 | -0.44 | -0.41 |
| HSD17B14 | -0.44 | -0.43 |
| AL138743.2 | -0.44 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity | TAS | glutamate (GO term level: 6) | 10198162 |
| GO:0005529 | sugar binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016051 | carbohydrate biosynthetic process | IEA | - | |
| GO:0006112 | energy reserve metabolic process | TAS | 10198162 | |
| GO:0006002 | fructose 6-phosphate metabolic process | TAS | 10198162 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0006541 | glutamine metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALANINE ASPARTATE AND GLUTAMATE METABOLISM | 32 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | 14 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME ASPARAGINE N LINKED GLYCOSYLATION | 81 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | 29 | 17 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING VIA NFKB | 28 | 21 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 2 TRANSIENTLY INDUCED BY EGF | 51 | 29 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 136 | 142 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 287 | 293 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-143 | 469 | 476 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-153 | 830 | 837 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-219 | 817 | 823 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-25/32/92/363/367 | 256 | 263 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 287 | 294 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 217 | 224 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-448 | 831 | 837 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.