Gene Page: RBX1
Summary ?
| GeneID | 9978 |
| Symbol | RBX1 |
| Synonyms | BA554C12.1|RNF75|ROC1 |
| Description | ring-box 1, E3 ubiquitin protein ligase |
| Reference | MIM:603814|HGNC:HGNC:9928|HPRD:06794| |
| Gene type | protein-coding |
| Map location | 22q13.2 |
| Pascal p-value | 1.772E-6 |
| Sherlock p-value | 0.603 |
| Fetal beta | -0.098 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
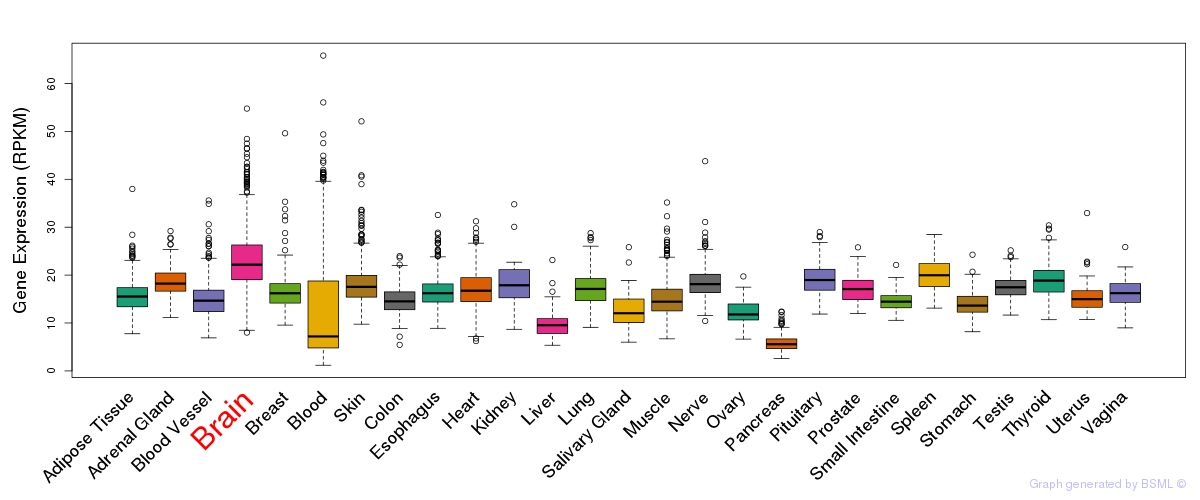
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AMBRA1 | 0.86 | 0.91 |
| FZR1 | 0.86 | 0.89 |
| LASP1 | 0.85 | 0.91 |
| SH3BP5L | 0.85 | 0.92 |
| STRN4 | 0.85 | 0.88 |
| GLT25D1 | 0.85 | 0.90 |
| FOXK2 | 0.85 | 0.92 |
| SARM1 | 0.84 | 0.91 |
| ITPKC | 0.84 | 0.86 |
| PPAPDC2 | 0.84 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.71 | -0.82 |
| MT-CO2 | -0.70 | -0.82 |
| AF347015.33 | -0.69 | -0.78 |
| AF347015.27 | -0.69 | -0.80 |
| FXYD1 | -0.69 | -0.78 |
| AF347015.8 | -0.68 | -0.82 |
| MT-CYB | -0.68 | -0.79 |
| COPZ2 | -0.68 | -0.76 |
| S100B | -0.67 | -0.76 |
| AC021016.1 | -0.67 | -0.79 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | Affinity Capture-MS | BioGRID | 12481031 |
| CAND1 | DKFZp434M1414 | FLJ10114 | FLJ10929 | FLJ38691 | FLJ90441 | KIAA0829 | TIP120 | TIP120A | cullin-associated and neddylation-dissociated 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12609982 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 11311237 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | Affinity Capture-Western | BioGRID | 11311237 |
| CCND3 | - | cyclin D3 | Affinity Capture-Western | BioGRID | 11311237 |
| CDC34 | E2-CDC34 | UBC3 | UBE2R1 | cell division cycle 34 homolog (S. cerevisiae) | Reconstituted Complex | BioGRID | 11675391 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | - | HPRD | 11337588 |
| CUL1 | MGC149834 | MGC149835 | cullin 1 | CUL1 interacts with ROC1. | BIND | 11956208 |
| CUL1 | MGC149834 | MGC149835 | cullin 1 | - | HPRD,BioGRID | 11961546 |
| CUL1 | MGC149834 | MGC149835 | cullin 1 | Rbx1 interacts with Cul1 [411-776] to form part of the Cul1-Rbx1-Skp1-F Boxskp2 Scf Ubiquitin Ligase complex. | BIND | 11961546 |
| CUL2 | MGC131970 | cullin 2 | Affinity Capture-MS | BioGRID | 12481031 |
| CUL3 | - | cullin 3 | - | HPRD | 11311237 |
| CUL3 | - | cullin 3 | Affinity Capture-MS | BioGRID | 12481031 |
| CUL4A | - | cullin 4A | ROC1 interacts with Cul4A. | BIND | 15448697 |
| CUL4A | - | cullin 4A | Affinity Capture-MS | BioGRID | 12481031 |
| CUL4B | DKFZp686F1470 | KIAA0695 | MRXHF2 | MRXSC | SFM2 | cullin 4B | Affinity Capture-MS | BioGRID | 12481031 |
| CUL5 | VACM-1 | VACM1 | cullin 5 | Rbx1 interacts with Cul5. | BIND | 14564014 |
| CUL7 | KIAA0076 | dJ20C7.5 | cullin 7 | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12481031 |
| DET1 | FLJ10103 | MGC126156 | MGC126157 | de-etiolated homolog 1 (Arabidopsis) | Affinity Capture-Western | BioGRID | 14739464 |
| FBXO18 | FBH1 | FLJ14590 | Fbx18 | MGC131916 | MGC141935 | MGC141937 | F-box protein, helicase, 18 | - | HPRD,BioGRID | 11956208 |
| FBXO22 | FBX22 | FISTC1 | FLJ13986 | MGC31799 | F-box protein 22 | Affinity Capture-MS | BioGRID | 12481031 |
| FBXO3 | DKFZp564B092 | FBA | FBX3 | F-box protein 3 | Affinity Capture-MS | BioGRID | 12481031 |
| FBXW8 | FBW6 | FBW8 | FBX29 | FBXO29 | FBXW6 | MGC33534 | F-box and WD repeat domain containing 8 | Affinity Capture-Western | BioGRID | 12481031 |
| GPS1 | COPS1 | CSN1 | MGC71287 | G protein pathway suppressor 1 | - | HPRD | 11337588 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | Affinity Capture-MS | BioGRID | 12481031 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Affinity Capture-MS | BioGRID | 12481031 |
| NEDD8 | FLJ43224 | MGC104393 | MGC125896 | MGC125897 | Nedd-8 | neural precursor cell expressed, developmentally down-regulated 8 | Affinity Capture-MS | BioGRID | 12481031 |
| RFWD2 | COP1 | FLJ10416 | RNF200 | ring finger and WD repeat domain 2 | Affinity Capture-Western | BioGRID | 14739464 |
| SKP1 | EMC19 | MGC34403 | OCP-II | OCP2 | SKP1A | TCEB1L | p19A | S-phase kinase-associated protein 1 | Affinity Capture-Western | BioGRID | 12628165 |
| TUBA1B | K-ALPHA-1 | tubulin, alpha 1b | Affinity Capture-MS | BioGRID | 12481031 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NUCLEOTIDE EXCISION REPAIR | 44 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P27 PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLACTIN RECEPTOR SIGNALING | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | 52 | 30 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 24HR 5 UP | 34 | 23 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |