Gene Page: ZZEF1
Summary ?
| GeneID | 23140 |
| Symbol | ZZEF1 |
| Synonyms | ZZZ4 |
| Description | zinc finger ZZ-type and EF-hand domain containing 1 |
| Reference | HGNC:HGNC:29027|Ensembl:ENSG00000074755|HPRD:15906|Vega:OTTHUMG00000090741 |
| Gene type | protein-coding |
| Map location | 17p13.2 |
| Pascal p-value | 0.042 |
| Sherlock p-value | 0.241 |
| Fetal beta | 0.474 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
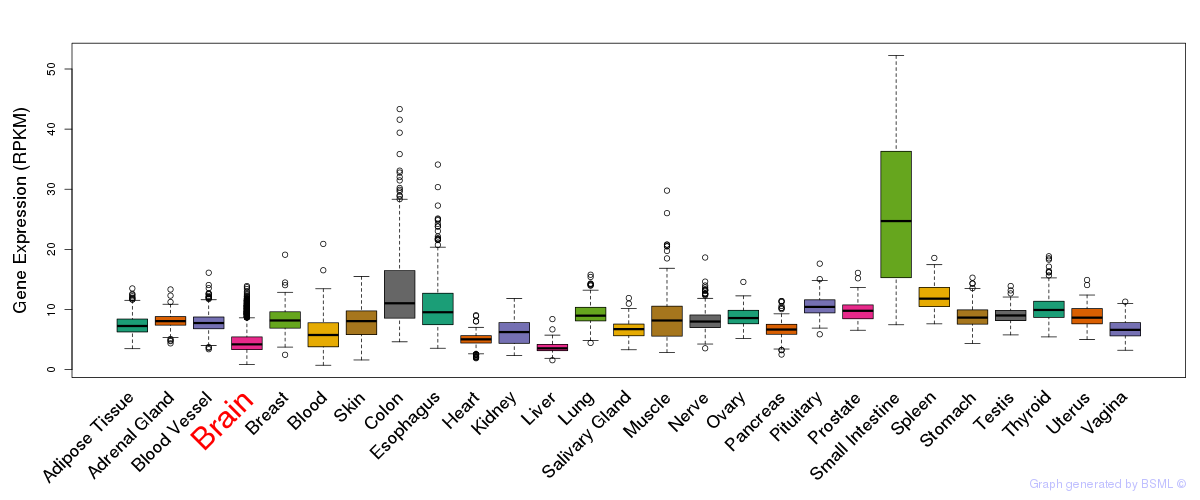
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TFG | 0.88 | 0.83 |
| PIGO | 0.87 | 0.82 |
| C7orf42 | 0.85 | 0.84 |
| GNS | 0.85 | 0.86 |
| GANAB | 0.85 | 0.84 |
| DPAGT1 | 0.85 | 0.82 |
| WDR45L | 0.84 | 0.76 |
| C16orf62 | 0.84 | 0.80 |
| GLB1 | 0.84 | 0.78 |
| VPS52 | 0.83 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.70 | -0.65 |
| MT-CO2 | -0.63 | -0.60 |
| AF347015.31 | -0.62 | -0.60 |
| AF347015.8 | -0.62 | -0.60 |
| AC098691.2 | -0.59 | -0.54 |
| AF347015.27 | -0.58 | -0.57 |
| AF347015.18 | -0.58 | -0.62 |
| AF347015.33 | -0.57 | -0.55 |
| MT-CYB | -0.57 | -0.56 |
| AF347015.2 | -0.56 | -0.56 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 17P13 DELETION | 21 | 15 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |