Gene Page: PRG4
Summary ?
| GeneID | 10216 |
| Symbol | PRG4 |
| Synonyms | CACP|HAPO|JCAP|MSF|SZP |
| Description | proteoglycan 4 |
| Reference | MIM:604283|HGNC:HGNC:9364|Ensembl:ENSG00000116690|HPRD:05047|Vega:OTTHUMG00000035574 |
| Gene type | protein-coding |
| Map location | 1q25-q31 |
| Fetal beta | -0.026 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs631503 | chr1 | 72327801 | PRG4 | 10216 | 0.08 | trans | ||
| rs2800921 | chr1 | 209368740 | PRG4 | 10216 | 0.03 | trans | ||
| rs7529880 | chr1 | 232598502 | PRG4 | 10216 | 0.03 | trans | ||
| rs12619086 | chr2 | 15180757 | PRG4 | 10216 | 0.1 | trans | ||
| rs17535450 | chr2 | 39900012 | PRG4 | 10216 | 0.05 | trans | ||
| rs17558168 | chr2 | 72151944 | PRG4 | 10216 | 0.05 | trans | ||
| rs16829215 | chr2 | 151871444 | PRG4 | 10216 | 0.1 | trans | ||
| rs6726013 | chr2 | 189064570 | PRG4 | 10216 | 0.1 | trans | ||
| rs4684042 | chr3 | 10559201 | PRG4 | 10216 | 0.19 | trans | ||
| rs1050604 | chr3 | 16357707 | PRG4 | 10216 | 0 | trans | ||
| rs11706314 | chr3 | 45309716 | PRG4 | 10216 | 0.19 | trans | ||
| rs9288980 | chr3 | 112964991 | PRG4 | 10216 | 0.14 | trans | ||
| rs3995908 | chr3 | 112998529 | PRG4 | 10216 | 0.14 | trans | ||
| rs7652801 | chr3 | 113008605 | PRG4 | 10216 | 0.14 | trans | ||
| rs6825572 | chr4 | 36215470 | PRG4 | 10216 | 0.13 | trans | ||
| rs17088743 | chr4 | 58668728 | PRG4 | 10216 | 0.1 | trans | ||
| rs17046025 | chr4 | 166066605 | PRG4 | 10216 | 0.11 | trans | ||
| rs1833564 | chr4 | 181732225 | PRG4 | 10216 | 0.05 | trans | ||
| rs416393 | chr5 | 9998327 | PRG4 | 10216 | 0.14 | trans | ||
| rs10041298 | chr5 | 52214802 | PRG4 | 10216 | 0.01 | trans | ||
| rs1462113 | chr5 | 152693290 | PRG4 | 10216 | 0.19 | trans | ||
| rs177521 | chr5 | 152767641 | PRG4 | 10216 | 0.04 | trans | ||
| rs545290 | chr5 | 152858437 | PRG4 | 10216 | 0.01 | trans | ||
| rs4236095 | chr6 | 47368686 | PRG4 | 10216 | 0.04 | trans | ||
| rs9451867 | chr6 | 92659609 | PRG4 | 10216 | 0 | trans | ||
| rs2301464 | chr6 | 112029983 | PRG4 | 10216 | 0.17 | trans | ||
| rs17704432 | chr6 | 112048867 | PRG4 | 10216 | 0.05 | trans | ||
| rs9374283 | chr6 | 112053416 | PRG4 | 10216 | 0.05 | trans | ||
| rs17072888 | chr6 | 112099533 | PRG4 | 10216 | 0.11 | trans | ||
| rs9321275 | chr6 | 131493217 | PRG4 | 10216 | 0.03 | trans | ||
| rs2398707 | chr7 | 2113428 | PRG4 | 10216 | 0 | trans | ||
| rs3778948 | chr7 | 2123290 | PRG4 | 10216 | 0.06 | trans | ||
| rs3778951 | chr7 | 2125319 | PRG4 | 10216 | 0 | trans | ||
| rs17157992 | chr7 | 83349706 | PRG4 | 10216 | 0.11 | trans | ||
| rs17215901 | chr8 | 106549085 | PRG4 | 10216 | 0.15 | trans | ||
| snp_a-2171708 | 0 | PRG4 | 10216 | 0.1 | trans | |||
| rs13279114 | chr8 | 106582168 | PRG4 | 10216 | 0.1 | trans | ||
| rs16908578 | chr8 | 139226337 | PRG4 | 10216 | 0.01 | trans | ||
| rs16908622 | chr8 | 139231604 | PRG4 | 10216 | 2.41E-4 | trans | ||
| rs1999431 | chr9 | 107564675 | PRG4 | 10216 | 0.13 | trans | ||
| rs998410 | chr9 | 117622673 | PRG4 | 10216 | 0 | trans | ||
| rs7929546 | chr11 | 8502946 | PRG4 | 10216 | 0.15 | trans | ||
| rs7102605 | chr11 | 8551676 | PRG4 | 10216 | 0.15 | trans | ||
| rs10437594 | chr11 | 8629423 | PRG4 | 10216 | 0.15 | trans | ||
| rs11042027 | chr11 | 8685450 | PRG4 | 10216 | 0.15 | trans | ||
| rs638035 | chr11 | 35509120 | PRG4 | 10216 | 0.17 | trans | ||
| rs11820030 | chr11 | 76133564 | PRG4 | 10216 | 0.01 | trans | ||
| rs17134872 | chr11 | 76139845 | PRG4 | 10216 | 0 | trans | ||
| rs10751255 | 0 | PRG4 | 10216 | 0.03 | trans | |||
| rs11236774 | chr11 | 76234952 | PRG4 | 10216 | 0 | trans | ||
| rs4759814 | chr12 | 131455220 | PRG4 | 10216 | 0.14 | trans | ||
| rs17055644 | chr13 | 38020671 | PRG4 | 10216 | 0.02 | trans | ||
| rs4349050 | chr13 | 41958722 | PRG4 | 10216 | 0.09 | trans | ||
| rs7982172 | chr13 | 41969697 | PRG4 | 10216 | 0.08 | trans | ||
| rs4942043 | chr13 | 41978218 | PRG4 | 10216 | 0.05 | trans | ||
| rs4402447 | chr13 | 41984741 | PRG4 | 10216 | 0.18 | trans | ||
| rs17057812 | chr13 | 60753449 | PRG4 | 10216 | 0 | trans | ||
| rs17251939 | chr14 | 49430822 | PRG4 | 10216 | 0.2 | trans | ||
| rs8061703 | chr16 | 81000281 | PRG4 | 10216 | 0.05 | trans | ||
| rs16941390 | chr17 | 45050984 | PRG4 | 10216 | 0.01 | trans | ||
| rs7236104 | chr18 | 14032818 | PRG4 | 10216 | 0.04 | trans | ||
| rs11873703 | chr18 | 61448702 | PRG4 | 10216 | 0.05 | trans | ||
| rs16967535 | chr19 | 33373697 | PRG4 | 10216 | 0.18 | trans | ||
| rs16967540 | chr19 | 33375874 | PRG4 | 10216 | 0.02 | trans | ||
| snp_a-1784374 | 0 | PRG4 | 10216 | 0.02 | trans | |||
| rs6510303 | chr19 | 33380831 | PRG4 | 10216 | 0.02 | trans | ||
| rs16993349 | chr20 | 7266857 | PRG4 | 10216 | 0.1 | trans | ||
| rs16993414 | chr20 | 7308537 | PRG4 | 10216 | 0.1 | trans | ||
| rs6095741 | chr20 | 48666589 | PRG4 | 10216 | 0.01 | trans | ||
| rs4822938 | chr22 | 28164049 | PRG4 | 10216 | 0.08 | trans | ||
| rs6000401 | chr22 | 37149335 | PRG4 | 10216 | 3.829E-4 | trans | ||
| rs2982104 | chrX | 94740630 | PRG4 | 10216 | 0.14 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
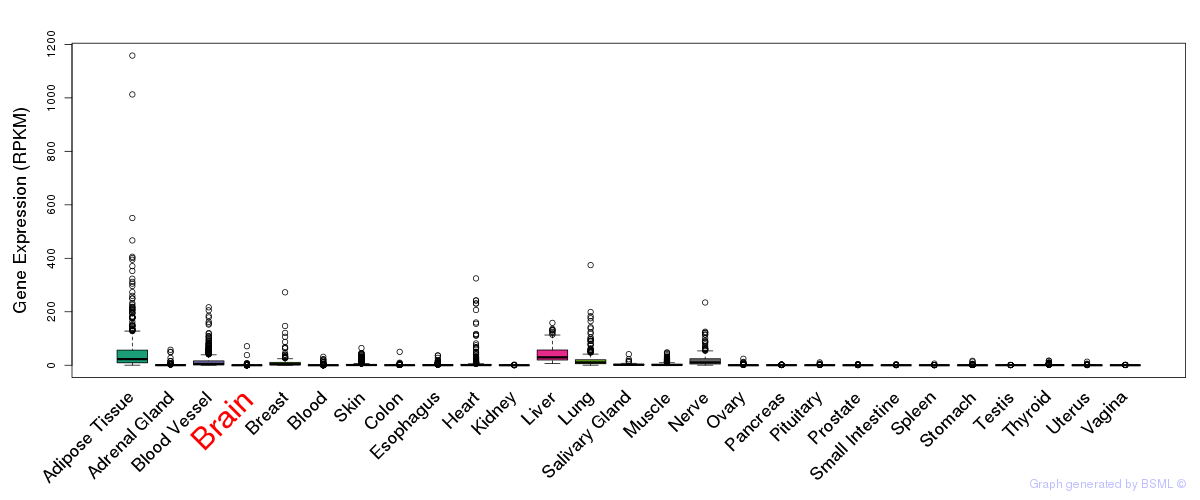
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR82 | 0.97 | 0.98 |
| CSDE1 | 0.97 | 0.97 |
| PHF20 | 0.96 | 0.97 |
| EPM2AIP1 | 0.96 | 0.97 |
| WDR48 | 0.96 | 0.97 |
| VEZT | 0.96 | 0.96 |
| EPC2 | 0.96 | 0.97 |
| POLR3A | 0.96 | 0.97 |
| ZMYM4 | 0.96 | 0.96 |
| EXOC2 | 0.96 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.79 | -0.89 |
| AF347015.31 | -0.79 | -0.88 |
| FXYD1 | -0.79 | -0.89 |
| IFI27 | -0.77 | -0.88 |
| AF347015.33 | -0.77 | -0.85 |
| HIGD1B | -0.76 | -0.88 |
| AF347015.27 | -0.76 | -0.85 |
| MT-CYB | -0.76 | -0.85 |
| AF347015.8 | -0.76 | -0.87 |
| TSC22D4 | -0.74 | -0.81 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MOROSETTI FACIOSCAPULOHUMERAL MUSCULAR DISTROPHY UP | 20 | 11 | All SZGR 2.0 genes in this pathway |
| KONG E2F1 TARGETS | 10 | 6 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CHEN LIVER METABOLISM QTL CIS | 93 | 40 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |