Gene Page: SLC25A17
Summary ?
| GeneID | 10478 |
| Symbol | SLC25A17 |
| Synonyms | PMP34 |
| Description | solute carrier family 25 member 17 |
| Reference | MIM:606795|HGNC:HGNC:10987|Ensembl:ENSG00000100372|HPRD:08425|Vega:OTTHUMG00000151139 |
| Gene type | protein-coding |
| Map location | 22q13.2 |
| Pascal p-value | 2.405E-4 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26548134 | 22 | 41185283 | SLC25A17 | 2.71E-5 | 0.437 | 0.018 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
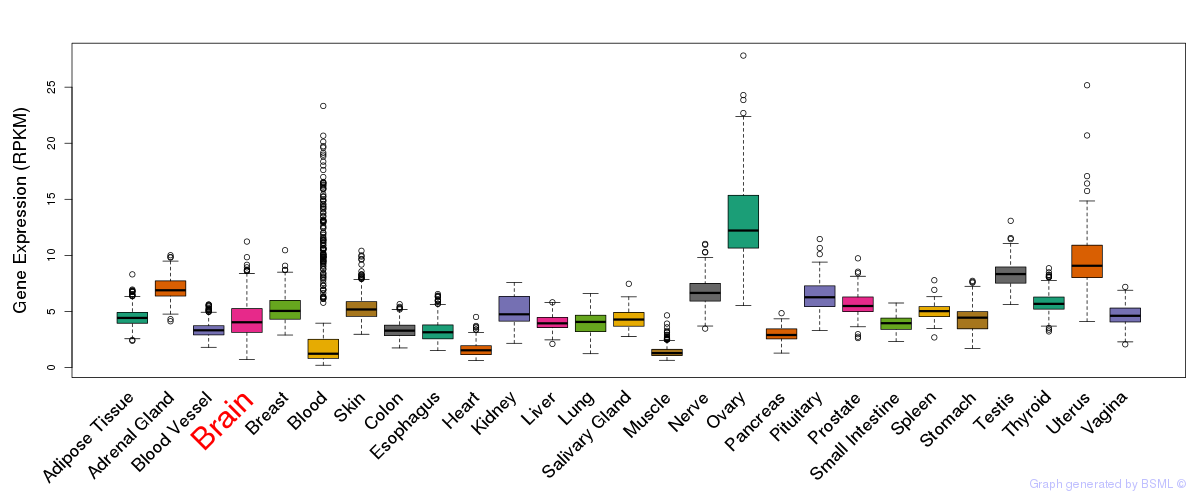
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SETDB1 | 0.93 | 0.93 |
| TMEM57 | 0.92 | 0.94 |
| HNRNPR | 0.92 | 0.94 |
| DHX29 | 0.92 | 0.94 |
| CNOT2 | 0.92 | 0.93 |
| RLF | 0.92 | 0.94 |
| MLLT3 | 0.91 | 0.94 |
| NCBP2 | 0.91 | 0.92 |
| KDM4A | 0.91 | 0.93 |
| FDXACB1 | 0.91 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AIFM3 | -0.77 | -0.82 |
| AF347015.31 | -0.76 | -0.88 |
| FXYD1 | -0.75 | -0.89 |
| TSC22D4 | -0.74 | -0.83 |
| MT-CO2 | -0.74 | -0.87 |
| AF347015.27 | -0.74 | -0.86 |
| AF347015.33 | -0.74 | -0.86 |
| ALDOC | -0.74 | -0.75 |
| HLA-F | -0.73 | -0.79 |
| S100B | -0.73 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10704444 | |
| GO:0005215 | transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006839 | mitochondrial transport | IEA | glutamate (GO term level: 6) | - |
| GO:0006810 | transport | TAS | 9874197 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005743 | mitochondrial inner membrane | IEA | - | |
| GO:0005777 | peroxisome | TAS | 9874197 | |
| GO:0005778 | peroxisomal membrane | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9874197 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME PEROXISOMAL LIPID METABOLISM | 21 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |