Gene Page: APPBP2
Summary ?
| GeneID | 10513 |
| Symbol | APPBP2 |
| Synonyms | APP-BP2|HS.84084|PAT1 |
| Description | amyloid beta precursor protein binding protein 2 |
| Reference | MIM:605324|HGNC:HGNC:622|Ensembl:ENSG00000062725|HPRD:05616|Vega:OTTHUMG00000180048 |
| Gene type | protein-coding |
| Map location | 17q23.2 |
| Pascal p-value | 0.112 |
| Sherlock p-value | 0.309 |
| Fetal beta | -0.232 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16614940 | 17 | 58602862 | APPBP2 | 7.85E-10 | -0.024 | 1.03E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1570589 | chr9 | 116874354 | APPBP2 | 10513 | 0.05 | trans |
Section II. Transcriptome annotation
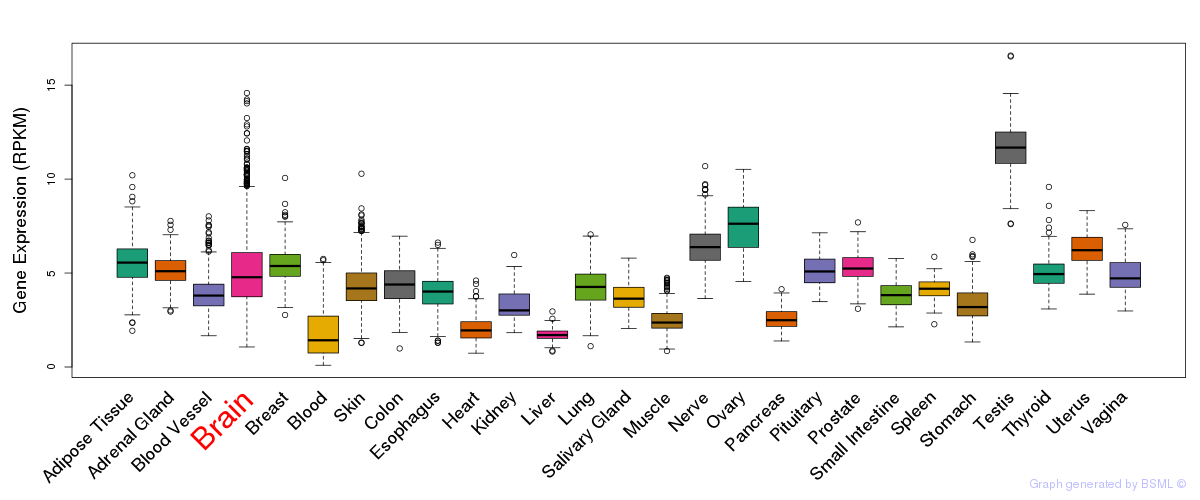
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD,BioGRID | 9843960 |
| APPBP2 | HS.84084 | KIAA0228 | PAT1 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | - | HPRD | 9843960 |
| CHMP5 | C9orf83 | CGI-34 | HSPC177 | PNAS-2 | SNF7DC2 | chromatin modifying protein 5 | Two-hybrid | BioGRID | 16189514 |
| CNTFR | MGC1774 | ciliary neurotrophic factor receptor | Two-hybrid | BioGRID | 16189514 |
| CREB3 | LUMAN | LZIP | MGC15333 | MGC19782 | cAMP responsive element binding protein 3 | Two-hybrid | BioGRID | 16189514 |
| EID3 | FLJ25832 | NSE4B | NSMCE4B | EP300 interacting inhibitor of differentiation 3 | Two-hybrid | BioGRID | 16189514 |
| KIF6 | C6orf102 | DKFZp451I2418 | MGC33317 | dJ1043E3.1 | dJ137F1.4 | dJ188D3.1 | kinesin family member 6 | Two-hybrid | BioGRID | 16189514 |
| MFSD3 | - | major facilitator superfamily domain containing 3 | Two-hybrid | BioGRID | 16189514 |
| MLLT3 | AF9 | FLJ2035 | YEATS3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | Two-hybrid | BioGRID | 16189514 |
| MUL1 | C1orf166 | FLJ12875 | GIDE | MAPL | MULAN | RNF218 | RP11-401M16.2 | mitochondrial ubiquitin ligase activator of NFKB 1 | Two-hybrid | BioGRID | 16189514 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | Two-hybrid | BioGRID | 16189514 |
| PCSK5 | PC5 | PC6 | PC6A | SPC6 | proprotein convertase subtilisin/kexin type 5 | Two-hybrid | BioGRID | 16189514 |
| RRS1 | KIAA0112 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| SLC22A6 | FLJ55736 | HOAT1 | MGC45260 | OAT1 | PAHT | ROAT1 | solute carrier family 22 (organic anion transporter), member 6 | Two-hybrid | BioGRID | 16189514 |
| SYT11 | DKFZp781D015 | KIAA0080 | MGC10881 | MGC17226 | SYT12 | synaptotagmin XI | Two-hybrid | BioGRID | 16189514 |
| TBL3 | SAZD | transducin (beta)-like 3 | Two-hybrid | BioGRID | 16189514 |
| WDR24 | C16orf21 | DKFZp434F054 | JFP7 | WD repeat domain 24 | Two-hybrid | BioGRID | 16189514 |
| WNT5B | MGC2648 | wingless-type MMTV integration site family, member 5B | Two-hybrid | BioGRID | 16189514 |
| ZCCHC12 | FLJ16123 | SIZN | SIZN1 | zinc finger, CCHC domain containing 12 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| NAM FXYD5 TARGETS DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO ANDROGEN UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO FORSKOLIN DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |