Gene Page: RALBP1
Summary ?
| GeneID | 10928 |
| Symbol | RALBP1 |
| Synonyms | RIP1|RLIP1|RLIP76 |
| Description | ralA binding protein 1 |
| Reference | MIM:605801|HGNC:HGNC:9841|Ensembl:ENSG00000017797|HPRD:09013|Vega:OTTHUMG00000131596 |
| Gene type | protein-coding |
| Map location | 18p11.3 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.011 |
| Fetal beta | -1.178 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18989081 | 18 | 9474805 | RALBP1 | 2.93E-9 | -0.009 | 2.05E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
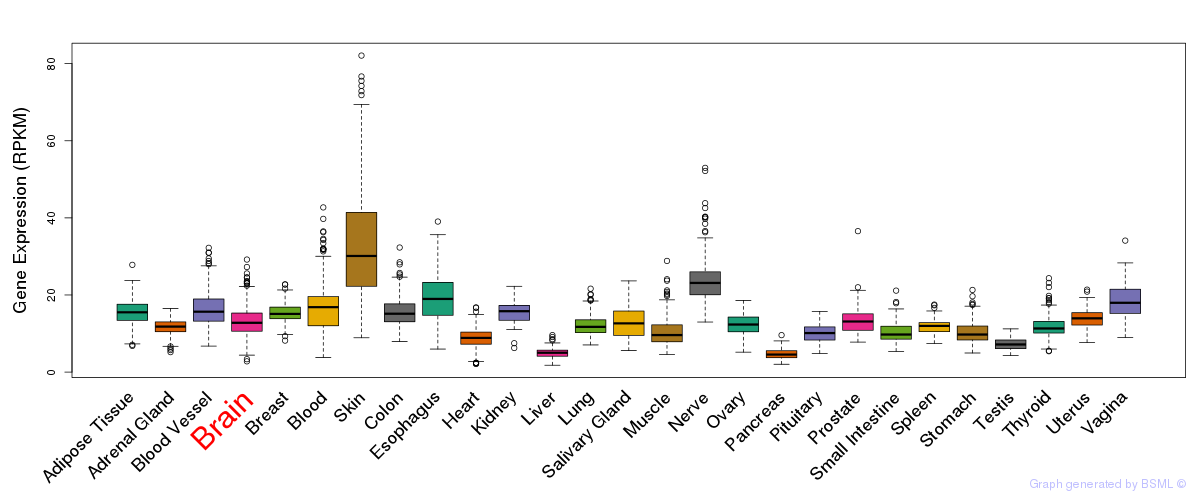
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACAN | 0.52 | 0.31 |
| SYT17 | 0.50 | 0.43 |
| FER1L6 | 0.48 | 0.37 |
| PTGER3 | 0.48 | 0.40 |
| ANKRD6 | 0.47 | 0.35 |
| MS4A8B | 0.46 | 0.28 |
| SGCZ | 0.46 | 0.27 |
| GRIK1 | 0.46 | 0.41 |
| SLIT3 | 0.45 | 0.37 |
| CX3CL1 | 0.45 | 0.34 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH2D2A | -0.26 | -0.28 |
| RPL14 | -0.25 | -0.24 |
| RPL18 | -0.25 | -0.28 |
| RPS19P3 | -0.25 | -0.25 |
| RPL19P12 | -0.24 | -0.25 |
| RPS13P2 | -0.24 | -0.24 |
| RPS23 | -0.24 | -0.24 |
| RPS6 | -0.24 | -0.26 |
| RPL28 | -0.24 | -0.29 |
| RPS16P1 | -0.24 | -0.29 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP2A2 | ADTAB | CLAPA2 | HIP9 | HYPJ | adaptor-related protein complex 2, alpha 2 subunit | RLIP76 interacts with alpha-adaptin. | BIND | 10910768 |
| AP2M1 | AP50 | CLAPM1 | mu2 | adaptor-related protein complex 2, mu 1 subunit | RLIP76 interacts with mu-2. | BIND | 10910768 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Affinity Capture-Western | BioGRID | 12887920 |
| CCNB1 | CCNB | cyclin B1 | - | HPRD,BioGRID | 12775724 |
| EPN1 | - | epsin 1 | Affinity Capture-Western | BioGRID | 12775724 |
| GTF3A | AP2 | TFIIIA | general transcription factor IIIA | Affinity Capture-Western | BioGRID | 12775724 |
| HOOK2 | FLJ26218 | HK2 | hook homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12621024 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | Affinity Capture-Western | BioGRID | 12621024 |
| NUMB | S171 | numb homolog (Drosophila) | Affinity Capture-Western | BioGRID | 12775724 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Rac1 interacts with RLIP76. | BIND | 7673236 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD,BioGRID | 7673236 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | - | HPRD,BioGRID | 7673236 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | RalA interacts with RalBP1. | BIND | 15592429 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | RalA interacts with RLIP76. | BIND | 7673236 |
| RALB | - | v-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein) | - | HPRD,BioGRID | 7673236 |
| RALB | - | v-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein) | RalB interacts with RLIP76. | BIND | 7673236 |
| REPS1 | RALBP1 | RALBP1 associated Eps domain containing 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9395447 |
| REPS2 | POB1 | RALBP1 associated Eps domain containing 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9422736 |
| SYNJ2BP | ARIP2 | FLJ11271 | FLJ41973 | OMP25 | synaptojanin 2 binding protein | Reconstituted Complex Two-hybrid | BioGRID | 11882656 |15451561 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | Affinity Capture-Western | BioGRID | 12887920 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | TNFR1 interacts with RIP1. | BIND | 15247912 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAC1 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PID RAC1 REG PATHWAY | 38 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LOPEZ TRANSLATION VIA FN1 SIGNALING | 35 | 21 | All SZGR 2.0 genes in this pathway |