Gene Page: CHGB
Summary ?
| GeneID | 1114 |
| Symbol | CHGB |
| Synonyms | SCG1 |
| Description | chromogranin B |
| Reference | MIM:118920|HGNC:HGNC:1930|Ensembl:ENSG00000089199|HPRD:07512|Vega:OTTHUMG00000031821 |
| Gene type | protein-coding |
| Map location | 20p12.3 |
| Pascal p-value | 0.558 |
| Sherlock p-value | 0.47 |
| Fetal beta | -1.661 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3747518 | chr2 | 163128724 | CHGB | 1114 | 0.17 | trans |
Section II. Transcriptome annotation
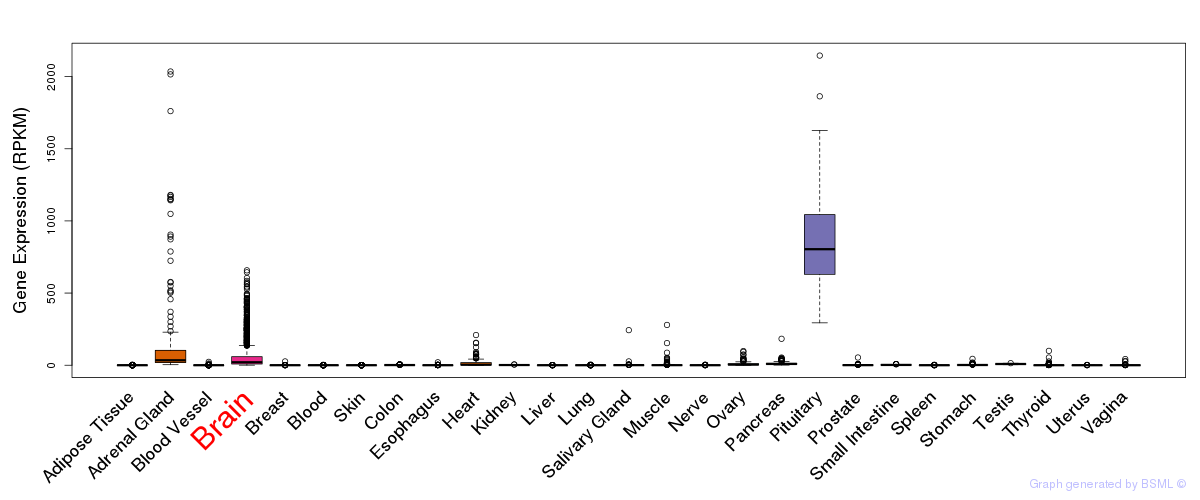
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRIM33 | 0.94 | 0.93 |
| IREB2 | 0.94 | 0.96 |
| RSBN1 | 0.93 | 0.95 |
| MED13 | 0.93 | 0.95 |
| ZNF148 | 0.93 | 0.96 |
| KIAA2022 | 0.93 | 0.96 |
| AFF4 | 0.93 | 0.95 |
| SLC38A1 | 0.93 | 0.96 |
| EPM2AIP1 | 0.93 | 0.96 |
| ZNF281 | 0.92 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.66 | -0.85 |
| TSC22D4 | -0.65 | -0.77 |
| AF347015.31 | -0.65 | -0.84 |
| HSD17B14 | -0.65 | -0.77 |
| HEPN1 | -0.65 | -0.73 |
| AIFM3 | -0.64 | -0.72 |
| MT-CO2 | -0.64 | -0.85 |
| ROM1 | -0.64 | -0.77 |
| S100B | -0.63 | -0.78 |
| SERPINB6 | -0.63 | -0.71 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005179 | hormone activity | TAS | 3608978 | |
| GO:0005515 | protein binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACACA | ACAC | ACC | ACC1 | ACCA | acetyl-Coenzyme A carboxylase alpha | Two-hybrid | BioGRID | 16169070 |
| ATXN2 | ATX2 | FLJ46772 | SCA2 | TNRC13 | ataxin 2 | Two-hybrid | BioGRID | 16169070 |
| CBFB | PEBP2B | core-binding factor, beta subunit | Two-hybrid | BioGRID | 16169070 |
| CCDC92 | FLJ22471 | coiled-coil domain containing 92 | Two-hybrid | BioGRID | 16169070 |
| FGFR3 | ACH | CD333 | CEK2 | HSFGFR3EX | JTK4 | fibroblast growth factor receptor 3 | Two-hybrid | BioGRID | 16169070 |
| GCS1 | - | glucosidase I | Two-hybrid | BioGRID | 16169070 |
| MARK3 | CTAK1 | KP78 | PAR1A | MAP/microtubule affinity-regulating kinase 3 | Two-hybrid | BioGRID | 16169070 |
| MGLL | HU-K5 | MGL | monoglyceride lipase | Two-hybrid | BioGRID | 16169070 |
| OGG1 | HMMH | HOGG1 | MUTM | OGH1 | 8-oxoguanine DNA glycosylase | Two-hybrid | BioGRID | 16169070 |
| POLD1 | CDC2 | POLD | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | Two-hybrid | BioGRID | 16169070 |
| PRAGMIN | DKFZp761P0423 | homolog of rat pragma of Rnd2 | Two-hybrid | BioGRID | 16169070 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | Two-hybrid | BioGRID | 16169070 |
| S100A8 | 60B8AG | CAGA | CFAG | CGLA | CP-10 | L1Ag | MA387 | MIF | MRP8 | NIF | P8 | S100 calcium binding protein A8 | Two-hybrid | BioGRID | 16169070 |
| SAFB2 | KIAA0138 | scaffold attachment factor B2 | Two-hybrid | BioGRID | 16169070 |
| SLC25A6 | AAC3 | ANT3 | ANT3Y | MGC17525 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | Two-hybrid | BioGRID | 16169070 |
| TAZ | BTHS | CMD3A | EFE | EFE2 | FLJ27390 | G4.5 | LVNCX | Taz1 | XAP-2 | tafazzin | Two-hybrid | BioGRID | 16169070 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | Two-hybrid | BioGRID | 16169070 |
| TWF2 | A6RP | A6r | MSTP011 | PTK9L | twinfilin, actin-binding protein, homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| YLPM1 | C14orf170 | ZAP | ZAP3 | YLP motif containing 1 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER F | 54 | 32 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP | 66 | 42 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO PROXIMAL DENDRITES | 37 | 26 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS UP | 46 | 30 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-544 | 94 | 100 | 1A | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.