Gene Page: CHRM4
Summary ?
| GeneID | 1132 |
| Symbol | CHRM4 |
| Synonyms | HM4|M4R |
| Description | cholinergic receptor muscarinic 4 |
| Reference | MIM:118495|HGNC:HGNC:1953|Ensembl:ENSG00000180720|HPRD:00330|Vega:OTTHUMG00000154371 |
| Gene type | protein-coding |
| Map location | 11p12-p11.2 |
| Pascal p-value | 3.478E-10 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
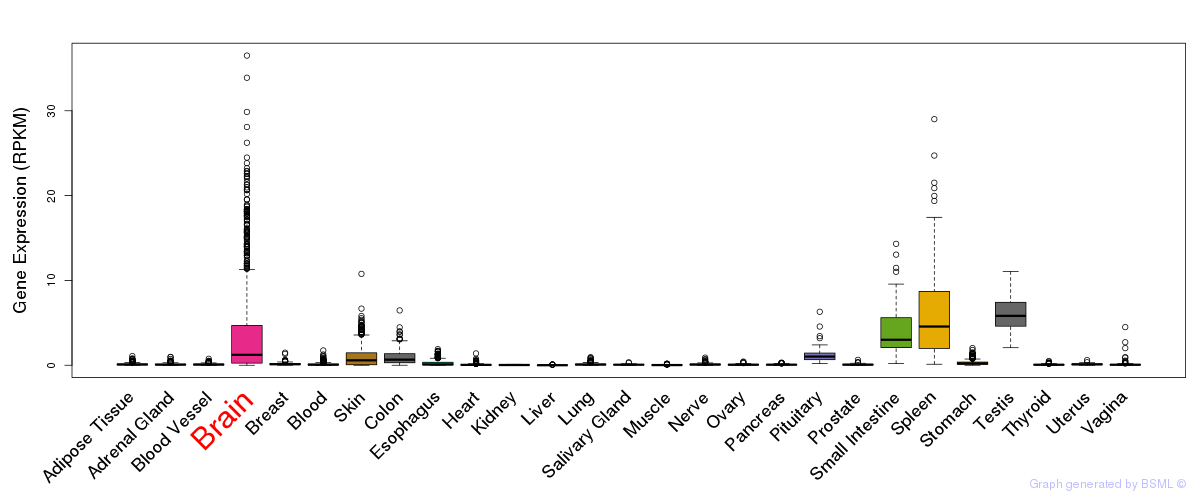
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STK19 | 0.83 | 0.79 |
| EVL | 0.82 | 0.79 |
| FBL | 0.81 | 0.78 |
| ZMYND19 | 0.80 | 0.77 |
| GNB2 | 0.79 | 0.78 |
| SCMH1 | 0.79 | 0.80 |
| KLHL22 | 0.79 | 0.77 |
| DTX1 | 0.78 | 0.77 |
| ADPRHL2 | 0.78 | 0.75 |
| MVD | 0.78 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.62 | -0.65 |
| AF347015.27 | -0.61 | -0.65 |
| AF347015.8 | -0.60 | -0.65 |
| AF347015.31 | -0.60 | -0.63 |
| AF347015.21 | -0.60 | -0.67 |
| MT-CYB | -0.59 | -0.63 |
| AF347015.33 | -0.58 | -0.62 |
| NOSTRIN | -0.55 | -0.59 |
| AF347015.15 | -0.55 | -0.62 |
| AF347015.2 | -0.54 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004981 | muscarinic acetylcholine receptor activity | TAS | 9603968 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007213 | acetylcholine receptor signaling, muscarinic pathway | TAS | 3443095 | |
| GO:0008283 | cell proliferation | TAS | 2739737 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | TAS | 3443095 | |
| GO:0005887 | integral to plasma membrane | TAS | 3037705 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC UP | 62 | 38 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |