Gene Page: CCR6
Summary ?
| GeneID | 1235 |
| Symbol | CCR6 |
| Synonyms | BN-1|C-C CKR-6|CC-CKR-6|CCR-6|CD196|CKR-L3|CKRL3|CMKBR6|DCR2|DRY6|GPR29|GPRCY4|STRL22 |
| Description | C-C motif chemokine receptor 6 |
| Reference | MIM:601835|HGNC:HGNC:1607|Ensembl:ENSG00000112486|HPRD:03498|Vega:OTTHUMG00000016015 |
| Gene type | protein-coding |
| Map location | 6q27 |
| Pascal p-value | 0.284 |
| Fetal beta | -1.118 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 2 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs3093024 | 6 | 167532793 | null | 1.499 | CCR6 | null | |
| rs3093023 | 6 | 167534290 | null | 1.489 | CCR6 | null |
Section II. Transcriptome annotation
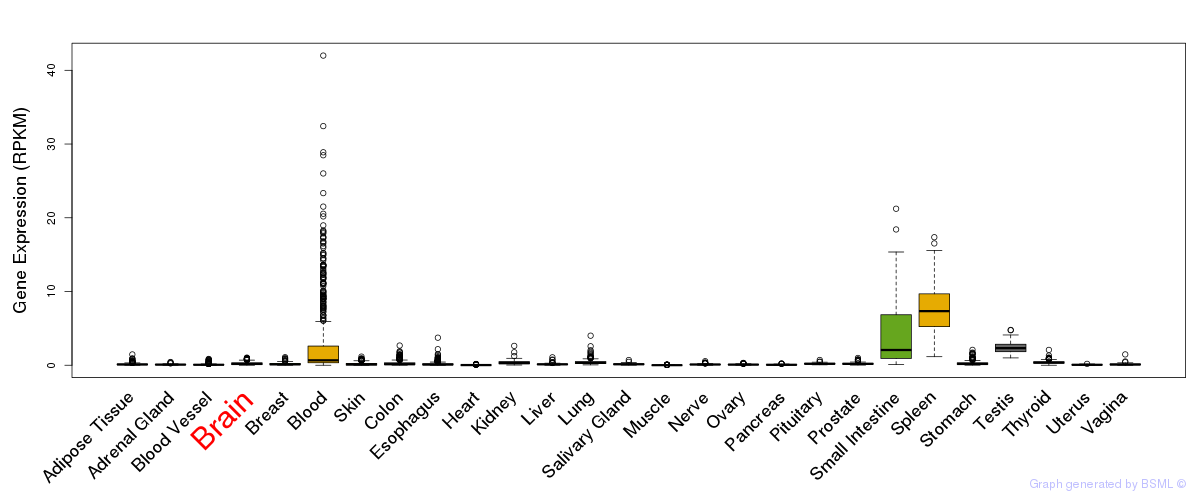
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STARD9 | 0.54 | 0.54 |
| RAD52 | 0.51 | 0.53 |
| PABPC1L | 0.51 | 0.45 |
| PAN2 | 0.51 | 0.54 |
| CEP152 | 0.50 | 0.50 |
| RTTN | 0.50 | 0.48 |
| GUSB | 0.50 | 0.50 |
| FANCA | 0.50 | 0.46 |
| FSCN3 | 0.50 | 0.40 |
| MYO9B | 0.50 | 0.53 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.32 | -0.36 |
| IL32 | -0.31 | -0.41 |
| MYL3 | -0.31 | -0.36 |
| IFI27 | -0.30 | -0.34 |
| MT-CO2 | -0.30 | -0.33 |
| GNG11 | -0.29 | -0.34 |
| CXCL14 | -0.29 | -0.35 |
| RERGL | -0.29 | -0.41 |
| HIGD1B | -0.29 | -0.34 |
| AF347015.21 | -0.28 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 9186513 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004945 | angiotensin type II receptor activity | IEA | - | |
| GO:0016493 | C-C chemokine receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007204 | elevation of cytosolic calcium ion concentration | TAS | 9223454 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0006968 | cellular defense response | TAS | 10521347 | |
| GO:0007165 | signal transduction | TAS | 9186513 | |
| GO:0006959 | humoral immune response | TAS | 11001880 | |
| GO:0006935 | chemotaxis | TAS | 11001880 | |
| GO:0006928 | cell motion | TAS | 9186513 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | TAS | 9186513 | |
| GO:0005887 | integral to plasma membrane | TAS | 9186513 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEFENSINS | 51 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME BETA DEFENSINS | 42 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | 57 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 SIGNALING 2 | 12 | 8 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 TARGETS WITH STAT5 BINDING SITES T1 | 9 | 7 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 DN | 38 | 22 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS UP | 46 | 30 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR DN | 55 | 35 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |