Gene Page: ADH1B
Summary ?
| GeneID | 125 |
| Symbol | ADH1B |
| Synonyms | ADH2|HEL-S-117 |
| Description | alcohol dehydrogenase 1B (class I), beta polypeptide |
| Reference | MIM:103720|HGNC:HGNC:250|HPRD:00065| |
| Gene type | protein-coding |
| Map location | 4q23 |
| Pascal p-value | 0.445 |
| Fetal beta | -0.197 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16841037 | chr1 | 4316543 | ADH1B | 125 | 0.03 | trans | ||
| rs17775722 | chr9 | 26199256 | ADH1B | 125 | 0.14 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
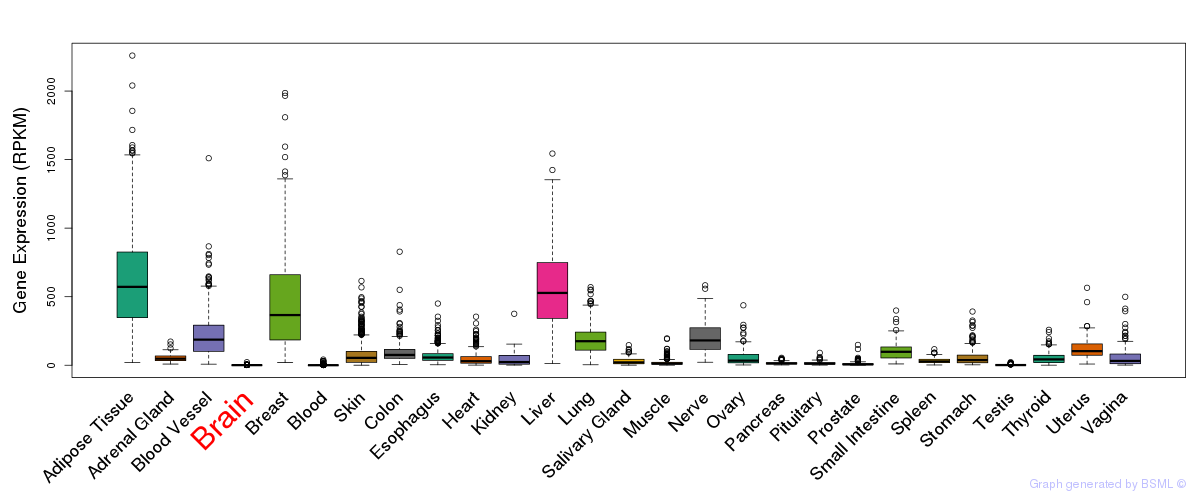
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HNRNPC | 0.93 | 0.89 |
| RBBP4 | 0.92 | 0.89 |
| GNAI3 | 0.91 | 0.81 |
| UBA2 | 0.91 | 0.87 |
| CPNE3 | 0.90 | 0.81 |
| ASF1A | 0.90 | 0.82 |
| CPSF3 | 0.90 | 0.85 |
| CLNS1A | 0.90 | 0.85 |
| MED28 | 0.90 | 0.86 |
| NPM1 | 0.89 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.33 | -0.59 | -0.74 |
| AF347015.27 | -0.58 | -0.71 |
| MT-CO2 | -0.58 | -0.71 |
| AF347015.8 | -0.58 | -0.72 |
| MT-CYB | -0.56 | -0.70 |
| AF347015.15 | -0.55 | -0.70 |
| AF347015.26 | -0.55 | -0.70 |
| SLC9A3R2 | -0.55 | -0.54 |
| TINAGL1 | -0.54 | -0.64 |
| HLA-F | -0.54 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004024 | alcohol dehydrogenase activity, zinc-dependent | TAS | 9982 | |
| GO:0005488 | binding | IEA | - | |
| GO:0008270 | zinc ion binding | IDA | 9982 | |
| GO:0009055 | electron carrier activity | TAS | 2398055 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006069 | ethanol oxidation | TAS | 2398055 | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG FATTY ACID METABOLISM | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG TYROSINE METABOLISM | 42 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG RETINOL METABOLISM | 64 | 37 | All SZGR 2.0 genes in this pathway |
| KEGG METABOLISM OF XENOBIOTICS BY CYTOCHROME P450 | 70 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM CYTOCHROME P450 | 72 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ETHANOL OXIDATION | 10 | 9 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR MARKERS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS DN | 57 | 33 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER | 49 | 29 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL OVERALL UP | 7 | 5 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL UP | 11 | 9 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR UP | 39 | 30 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |