Gene Page: ADH1C
Summary ?
| GeneID | 126 |
| Symbol | ADH1C |
| Synonyms | ADH3 |
| Description | alcohol dehydrogenase 1C (class I), gamma polypeptide |
| Reference | MIM:103730|HGNC:HGNC:251|Ensembl:ENSG00000248144|HPRD:00066|Vega:OTTHUMG00000161422 |
| Gene type | protein-coding |
| Map location | 4q23 |
| Pascal p-value | 0.218 |
| Fetal beta | -0.272 |
| eGene | Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1789892 | 4 | 100250658 | ADH1C | ENSG00000248144.1 | 9.324E-7 | 0.01 | 23526 | gtex_brain_putamen_basal |
| rs1154433 | 4 | 100253708 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 20476 | gtex_brain_putamen_basal |
| rs1229978 | 4 | 100256199 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 17985 | gtex_brain_putamen_basal |
| rs1662031 | 4 | 100256793 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 17391 | gtex_brain_putamen_basal |
| rs1612735 | 4 | 100258007 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 16177 | gtex_brain_putamen_basal |
| rs1789898 | 4 | 100258336 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 15848 | gtex_brain_putamen_basal |
| rs1442479 | 4 | 100258675 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 15509 | gtex_brain_putamen_basal |
| rs1442480 | 4 | 100258681 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 15503 | gtex_brain_putamen_basal |
| rs1789900 | 4 | 100258717 | ADH1C | ENSG00000248144.1 | 2.579E-6 | 0.01 | 15467 | gtex_brain_putamen_basal |
| rs1442481 | 4 | 100259047 | ADH1C | ENSG00000248144.1 | 1.013E-6 | 0.01 | 15137 | gtex_brain_putamen_basal |
| rs1693456 | 4 | 100259351 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 14833 | gtex_brain_putamen_basal |
| rs2851299 | 4 | 100259618 | ADH1C | ENSG00000248144.1 | 9.298E-7 | 0.01 | 14566 | gtex_brain_putamen_basal |
| rs1662060 | 4 | 100259841 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 14343 | gtex_brain_putamen_basal |
| rs1789902 | 4 | 100260171 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 14013 | gtex_brain_putamen_basal |
| rs1693476 | 4 | 100260274 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 13910 | gtex_brain_putamen_basal |
| rs1662059 | 4 | 100260378 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 13806 | gtex_brain_putamen_basal |
| rs698 | 4 | 100260789 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 13395 | gtex_brain_putamen_basal |
| rs2298755 | 4 | 100261038 | ADH1C | ENSG00000248144.1 | 9.759E-7 | 0.01 | 13146 | gtex_brain_putamen_basal |
| rs373899105 | 4 | 100261040 | ADH1C | ENSG00000248144.1 | 1.28E-6 | 0.01 | 13144 | gtex_brain_putamen_basal |
| rs3114048 | 4 | 100261216 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 12968 | gtex_brain_putamen_basal |
| rs199619002 | 4 | 100261576 | ADH1C | ENSG00000248144.1 | 8.334E-7 | 0.01 | 12608 | gtex_brain_putamen_basal |
| rs55753621 | 4 | 100261577 | ADH1C | ENSG00000248144.1 | 8.334E-7 | 0.01 | 12607 | gtex_brain_putamen_basal |
| rs1789903 | 4 | 100262041 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 12143 | gtex_brain_putamen_basal |
| rs1662058 | 4 | 100262759 | ADH1C | ENSG00000248144.1 | 9.017E-7 | 0.01 | 11425 | gtex_brain_putamen_basal |
| rs1693477 | 4 | 100263062 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 11122 | gtex_brain_putamen_basal |
| rs1789906 | 4 | 100263219 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10965 | gtex_brain_putamen_basal |
| rs904093 | 4 | 100263247 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10937 | gtex_brain_putamen_basal |
| rs904094 | 4 | 100263252 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10932 | gtex_brain_putamen_basal |
| rs904095 | 4 | 100263383 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10801 | gtex_brain_putamen_basal |
| rs1662057 | 4 | 100263414 | ADH1C | ENSG00000248144.1 | 8.875E-7 | 0.01 | 10770 | gtex_brain_putamen_basal |
| rs1789908 | 4 | 100263417 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10767 | gtex_brain_putamen_basal |
| rs904096 | 4 | 100263584 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10600 | gtex_brain_putamen_basal |
| rs1789910 | 4 | 100263715 | ADH1C | ENSG00000248144.1 | 8.989E-7 | 0.01 | 10469 | gtex_brain_putamen_basal |
| rs1693480 | 4 | 100263762 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10422 | gtex_brain_putamen_basal |
| rs1789911 | 4 | 100263778 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 10406 | gtex_brain_putamen_basal |
| rs1693481 | 4 | 100263834 | ADH1C | ENSG00000248144.1 | 9.759E-7 | 0.01 | 10350 | gtex_brain_putamen_basal |
| rs1789912 | 4 | 100263942 | ADH1C | ENSG00000248144.1 | 5.429E-7 | 0.01 | 10242 | gtex_brain_putamen_basal |
| rs1693482 | 4 | 100263965 | ADH1C | ENSG00000248144.1 | 8.774E-7 | 0.01 | 10219 | gtex_brain_putamen_basal |
| rs1631460 | 4 | 100264620 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 9564 | gtex_brain_putamen_basal |
| rs1693483 | 4 | 100264946 | ADH1C | ENSG00000248144.1 | 8.737E-7 | 0.01 | 9238 | gtex_brain_putamen_basal |
| rs1693424 | 4 | 100265236 | ADH1C | ENSG00000248144.1 | 9.022E-7 | 0.01 | 8948 | gtex_brain_putamen_basal |
| rs1625439 | 4 | 100265325 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 8859 | gtex_brain_putamen_basal |
| rs1789913 | 4 | 100265348 | ADH1C | ENSG00000248144.1 | 8.976E-7 | 0.01 | 8836 | gtex_brain_putamen_basal |
| rs62307298 | 4 | 100265550 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 8634 | gtex_brain_putamen_basal |
| rs1693425 | 4 | 100266112 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 8072 | gtex_brain_putamen_basal |
| rs1693426 | 4 | 100266330 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 7854 | gtex_brain_putamen_basal |
| rs1662053 | 4 | 100266583 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 7601 | gtex_brain_putamen_basal |
| rs1662052 | 4 | 100266635 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 7549 | gtex_brain_putamen_basal |
| rs1789916 | 4 | 100266696 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 7488 | gtex_brain_putamen_basal |
| rs1693427 | 4 | 100266827 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 7357 | gtex_brain_putamen_basal |
| rs1693428 | 4 | 100266931 | ADH1C | ENSG00000248144.1 | 1.096E-6 | 0.01 | 7253 | gtex_brain_putamen_basal |
| rs1789917 | 4 | 100267164 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 7020 | gtex_brain_putamen_basal |
| rs1662051 | 4 | 100267239 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 6945 | gtex_brain_putamen_basal |
| rs1789919 | 4 | 100267719 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 6465 | gtex_brain_putamen_basal |
| rs1693431 | 4 | 100268049 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 6135 | gtex_brain_putamen_basal |
| rs3762896 | 4 | 100268131 | ADH1C | ENSG00000248144.1 | 2.656E-6 | 0.01 | 6053 | gtex_brain_putamen_basal |
| rs4147542 | 4 | 100268553 | ADH1C | ENSG00000248144.1 | 2.656E-6 | 0.01 | 5631 | gtex_brain_putamen_basal |
| rs35500540 | 4 | 100268753 | ADH1C | ENSG00000248144.1 | 1.422E-6 | 0.01 | 5431 | gtex_brain_putamen_basal |
| rs77595642 | 4 | 100269259 | ADH1C | ENSG00000248144.1 | 1.473E-6 | 0.01 | 4925 | gtex_brain_putamen_basal |
| rs35081954 | 4 | 100270818 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 3366 | gtex_brain_putamen_basal |
| rs35920708 | 4 | 100271008 | ADH1C | ENSG00000248144.1 | 9.331E-7 | 0.01 | 3176 | gtex_brain_putamen_basal |
| rs1629270 | 4 | 100271306 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 2878 | gtex_brain_putamen_basal |
| rs1662049 | 4 | 100271849 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | 2335 | gtex_brain_putamen_basal |
| rs1662048 | 4 | 100272032 | ADH1C | ENSG00000248144.1 | 9.214E-7 | 0.01 | 2152 | gtex_brain_putamen_basal |
| rs6846835 | 4 | 100272147 | ADH1C | ENSG00000248144.1 | 2.642E-6 | 0.01 | 2037 | gtex_brain_putamen_basal |
| rs11936869 | 4 | 100273173 | ADH1C | ENSG00000248144.1 | 2.656E-6 | 0.01 | 1011 | gtex_brain_putamen_basal |
| rs67512205 | 4 | 100273368 | ADH1C | ENSG00000248144.1 | 1.751E-6 | 0.01 | 816 | gtex_brain_putamen_basal |
| rs4147541 | 4 | 100274157 | ADH1C | ENSG00000248144.1 | 2.656E-6 | 0.01 | 27 | gtex_brain_putamen_basal |
| rs1789924 | 4 | 100274286 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -102 | gtex_brain_putamen_basal |
| rs1629838 | 4 | 100274896 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -712 | gtex_brain_putamen_basal |
| rs11499824 | 4 | 100274983 | ADH1C | ENSG00000248144.1 | 2.666E-6 | 0.01 | -799 | gtex_brain_putamen_basal |
| rs2584453 | 4 | 100275305 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -1121 | gtex_brain_putamen_basal |
| rs1789925 | 4 | 100275745 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -1561 | gtex_brain_putamen_basal |
| rs1693469 | 4 | 100275902 | ADH1C | ENSG00000248144.1 | 1.993E-6 | 0.01 | -1718 | gtex_brain_putamen_basal |
| rs11499826 | 4 | 100276936 | ADH1C | ENSG00000248144.1 | 1.992E-7 | 0.01 | -2752 | gtex_brain_putamen_basal |
| rs10006545 | 4 | 100277432 | ADH1C | ENSG00000248144.1 | 2.656E-6 | 0.01 | -3248 | gtex_brain_putamen_basal |
| rs2453980 | 4 | 100277437 | ADH1C | ENSG00000248144.1 | 2.039E-6 | 0.01 | -3253 | gtex_brain_putamen_basal |
| rs201720605 | 4 | 100278004 | ADH1C | ENSG00000248144.1 | 9.698E-7 | 0.01 | -3820 | gtex_brain_putamen_basal |
| rs980972 | 4 | 100279247 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -5063 | gtex_brain_putamen_basal |
| rs1693470 | 4 | 100279489 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -5305 | gtex_brain_putamen_basal |
| rs1662039 | 4 | 100279684 | ADH1C | ENSG00000248144.1 | 8.964E-7 | 0.01 | -5500 | gtex_brain_putamen_basal |
| rs2851300 | 4 | 100279824 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -5640 | gtex_brain_putamen_basal |
| rs1234584 | 4 | 100280041 | ADH1C | ENSG00000248144.1 | 9.112E-7 | 0.01 | -5857 | gtex_brain_putamen_basal |
| rs1662021 | 4 | 100280107 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -5923 | gtex_brain_putamen_basal |
| rs1662020 | 4 | 100280110 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -5926 | gtex_brain_putamen_basal |
| rs1235415 | 4 | 100281159 | ADH1C | ENSG00000248144.1 | 8.93E-7 | 0.01 | -6975 | gtex_brain_putamen_basal |
| rs1229858 | 4 | 100281634 | ADH1C | ENSG00000248144.1 | 7.815E-7 | 0.01 | -7450 | gtex_brain_putamen_basal |
| rs1229857 | 4 | 100281708 | ADH1C | ENSG00000248144.1 | 8.72E-7 | 0.01 | -7524 | gtex_brain_putamen_basal |
| rs10031168 | 4 | 100281893 | ADH1C | ENSG00000248144.1 | 2.656E-6 | 0.01 | -7709 | gtex_brain_putamen_basal |
| rs1229856 | 4 | 100281905 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -7721 | gtex_brain_putamen_basal |
| rs1229855 | 4 | 100282295 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -8111 | gtex_brain_putamen_basal |
| rs1229854 | 4 | 100282550 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -8366 | gtex_brain_putamen_basal |
| rs1229851 | 4 | 100283825 | ADH1C | ENSG00000248144.1 | 9.033E-7 | 0.01 | -9641 | gtex_brain_putamen_basal |
| rs1154435 | 4 | 100285148 | ADH1C | ENSG00000248144.1 | 2.221E-6 | 0.01 | -10964 | gtex_brain_putamen_basal |
| rs1154438 | 4 | 100285887 | ADH1C | ENSG00000248144.1 | 1.668E-6 | 0.01 | -11703 | gtex_brain_putamen_basal |
| rs1154439 | 4 | 100286233 | ADH1C | ENSG00000248144.1 | 6.081E-7 | 0.01 | -12049 | gtex_brain_putamen_basal |
| rs1154440 | 4 | 100286447 | ADH1C | ENSG00000248144.1 | 9.005E-7 | 0.01 | -12263 | gtex_brain_putamen_basal |
| rs1229973 | 4 | 100287178 | ADH1C | ENSG00000248144.1 | 9.314E-7 | 0.01 | -12994 | gtex_brain_putamen_basal |
| rs1583974 | 4 | 100287812 | ADH1C | ENSG00000248144.1 | 9.671E-7 | 0.01 | -13628 | gtex_brain_putamen_basal |
| rs1154442 | 4 | 100287849 | ADH1C | ENSG00000248144.1 | 1.42E-6 | 0.01 | -13665 | gtex_brain_putamen_basal |
| rs1154444 | 4 | 100288089 | ADH1C | ENSG00000248144.1 | 9.9E-7 | 0.01 | -13905 | gtex_brain_putamen_basal |
| rs1154445 | 4 | 100288521 | ADH1C | ENSG00000248144.1 | 1.033E-6 | 0.01 | -14337 | gtex_brain_putamen_basal |
| rs1154447 | 4 | 100289034 | ADH1C | ENSG00000248144.1 | 1.089E-6 | 0.01 | -14850 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
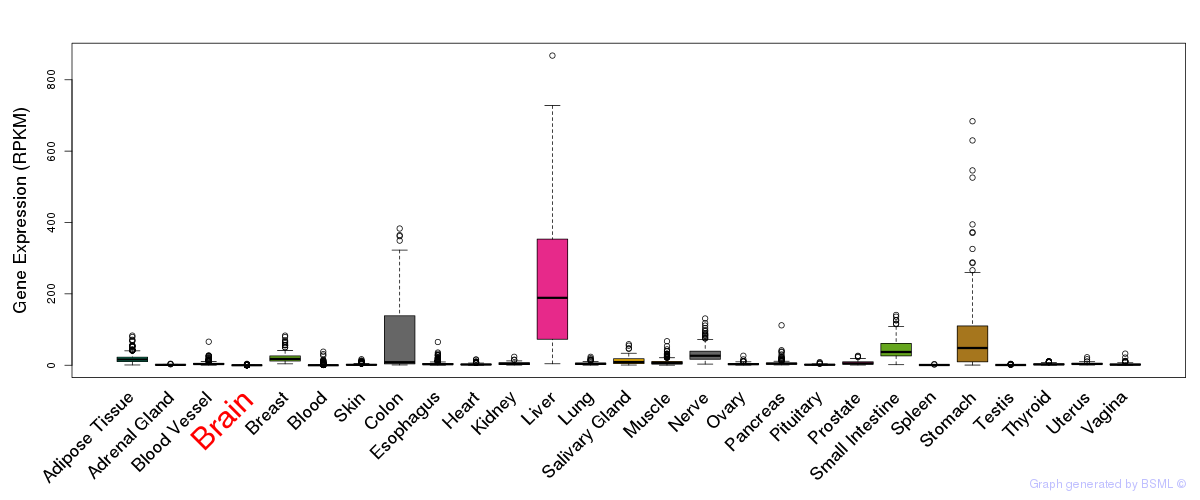
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG FATTY ACID METABOLISM | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG TYROSINE METABOLISM | 42 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG RETINOL METABOLISM | 64 | 37 | All SZGR 2.0 genes in this pathway |
| KEGG METABOLISM OF XENOBIOTICS BY CYTOCHROME P450 | 70 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM CYTOCHROME P450 | 72 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ETHANOL OXIDATION | 10 | 9 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 6 | 84 | 54 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN | 48 | 33 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS UP | 88 | 47 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR UP | 39 | 30 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |