Gene Page: DAPK1
Summary ?
| GeneID | 1612 |
| Symbol | DAPK1 |
| Synonyms | DAPK |
| Description | death-associated protein kinase 1 |
| Reference | MIM:600831|HGNC:HGNC:2674|Ensembl:ENSG00000196730|HPRD:02902|Vega:OTTHUMG00000020150 |
| Gene type | protein-coding |
| Map location | 9q21.33 |
| Pascal p-value | 0.4 |
| Sherlock p-value | 0.015 |
| Fetal beta | 0.506 |
| DMG | 2 (# studies) |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DAPK1 | chr9 | 90301641 | C | T | NM_004938 | p.N800N | synonymous SNV | Schizophrenia | DNM:McCarthy_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08797471 | 9 | 90113120 | DAPK1 | -0.024 | 0.36 | DMG:Nishioka_2013 | |
| cg19734228 | 9 | 90113593 | DAPK1 | 1.37E-9 | -0.018 | 1.37E-6 | DMG:Jaffe_2016 |
| cg08797471 | 9 | 90113120 | DAPK1 | 8.78E-9 | -0.022 | 4.04E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
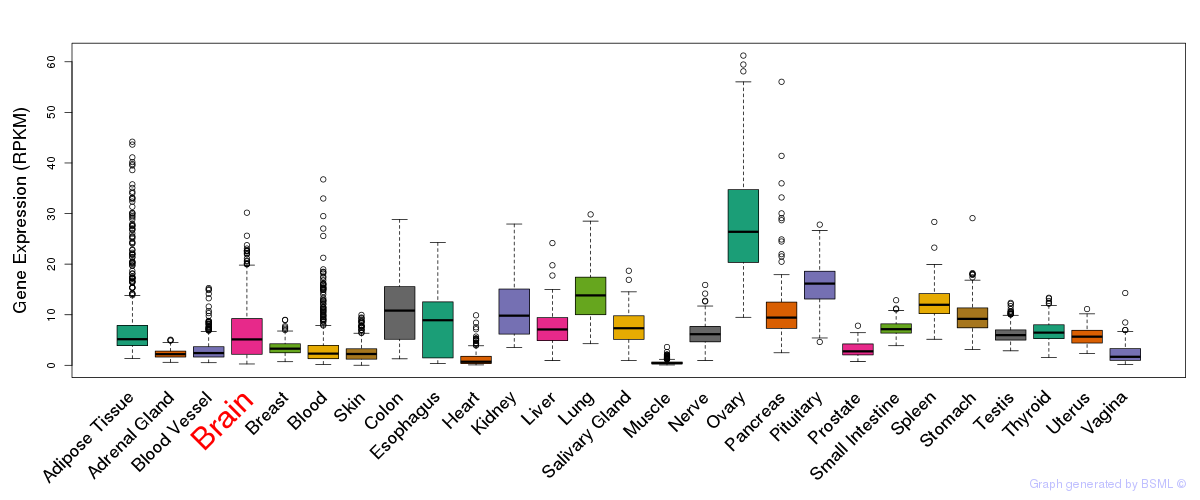
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD,BioGRID | 12911633 |
| KRIT1 | CAM | CCM1 | KRIT1, ankyrin repeat containing | Affinity Capture-Western | BioGRID | 12911633 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | DAPK interacts with Erk2. | BIND | 15616583 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | DAPK interacts with Erk1. | BIND | 15616583 |
| MIB1 | DIP-1 | DKFZp686I0769 | DKFZp761M1710 | FLJ90676 | MGC129659 | MGC129660 | MIB | ZZANK2 | ZZZ6 | mindbomb homolog 1 (Drosophila) | - | HPRD | 12351649 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 12911633 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 12911633 |
| UNC5A | FLJ16449 | KIAA1976 | UNC5H1 | unc-5 homolog A (C. elegans) | UNC5A (UNC5H1) interacts with DAPK1 (DAPK). This interaction was modelled on a demonstrated interaction between rat UNC5H1 and human DAPK. | BIND | 15729359 |
| UNC5B | UNC5H2 | p53RDL1 | unc-5 homolog B (C. elegans) | UNC5B (UNC5H2) interacts with DAPK1 (DAPK). This interaction was modelled on a demonstrated interaction between rat UNC5H2 and human DAPK. | BIND | 15729359 |
| UNC5C | UNC5H3 | unc-5 homolog C (C. elegans) | UNC5C (UNC5H3) interacts with DAPK1 (DAPK). This interaction was modelled on a demonstrated interaction between rat UNC5H3 and human DAPK. | BIND | 15729359 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD | 12911633 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-Western | BioGRID | 12911633 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF APOPTOSIS | 58 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| RAMJAUN APOPTOSIS BY TGFB1 VIA SMAD4 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER UP | 40 | 27 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| OHM EMBRYONIC CARCINOMA UP | 6 | 5 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 CORRELATED | 11 | 9 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| RADMACHER AML PROGNOSIS | 78 | 52 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| MCMURRAY TP53 HRAS COOPERATION RESPONSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER EARLY RECURRENCE UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 1HR UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 DN | 26 | 14 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE ALL DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |