Gene Page: DCTN1
Summary ?
| GeneID | 1639 |
| Symbol | DCTN1 |
| Synonyms | DAP-150|DP-150|P135 |
| Description | dynactin subunit 1 |
| Reference | MIM:601143|HGNC:HGNC:2711|Ensembl:ENSG00000204843|HPRD:07206|Vega:OTTHUMG00000129963 |
| Gene type | protein-coding |
| Map location | 2p13 |
| Pascal p-value | 0.543 |
| Sherlock p-value | 0.31 |
| Fetal beta | -1.376 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR TRAFFICKING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3203 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7598660 | chr2 | 73740424 | DCTN1 | 1639 | 0.13 | cis | ||
| rs11688718 | chr2 | 73760938 | DCTN1 | 1639 | 0.17 | cis | ||
| rs4852939 | chr2 | 73856768 | DCTN1 | 1639 | 0.08 | cis | ||
| rs4852316 | chr2 | 73919657 | DCTN1 | 1639 | 0.05 | cis | ||
| rs17350056 | chr2 | 73922876 | DCTN1 | 1639 | 0.15 | cis | ||
| rs12052539 | chr2 | 73937152 | DCTN1 | 1639 | 0.11 | cis | ||
| rs17350188 | chr2 | 73964841 | DCTN1 | 1639 | 0.09 | cis | ||
| rs12624267 | chr2 | 73986930 | DCTN1 | 1639 | 0.01 | cis |
Section II. Transcriptome annotation
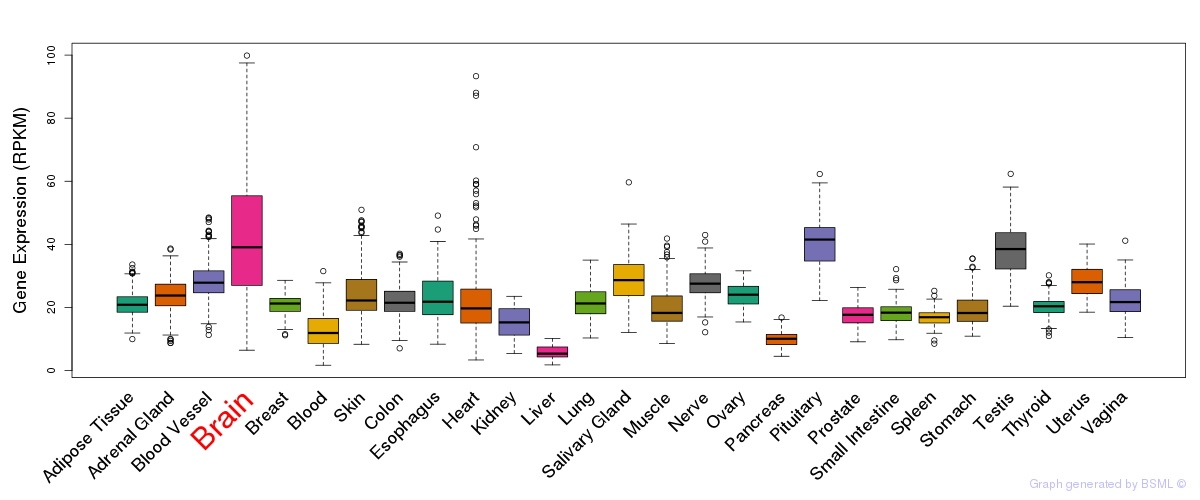
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003774 | motor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 9722614 |15107855 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | NAS | neurite (GO term level: 5) | 17360970 |
| GO:0007067 | mitosis | NAS | 1828535 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000922 | spindle pole | IDA | 14718566 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 1828535 | |
| GO:0030286 | dynein complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTR1A | ARP1 | CTRN1 | FLJ52695 | FLJ52800 | FLJ55002 | ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) | - | HPRD | 7878030 |
| AKTIP | FT1 | FTS | AKT interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | - | HPRD | 9361024 |
| BBS4 | - | Bardet-Biedl syndrome 4 | Affinity Capture-Western Two-hybrid | BioGRID | 15107855 |
| BICD2 | KIAA0699 | bA526D8.1 | bicaudal D homolog 2 (Drosophila) | - | HPRD | 11483508 |12447383 |
| BZW1 | BZAP45 | KIAA0005 | Nbla10236 | basic leucine zipper and W2 domains 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | - | HPRD | 11425872 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD | 11425872 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | - | HPRD | 11425872 |
| CLIP1 | CLIP | CLIP-170 | CLIP170 | CYLN1 | MGC131604 | RSN | CAP-GLY domain containing linker protein 1 | - | HPRD | 12789661 |
| DCTN2 | DCTN50 | DYNAMITIN | RBP50 | dynactin 2 (p50) | p50dynamitin (DCTN2) interacts with an unspecified isoform of p150glued (DCTN1). | BIND | 15580264 |
| DST | BP240 | BPA | BPAG1 | CATX-15 | D6S1101 | DKFZp564B2416 | DMH | DT | FLJ46791 | KIAA0465 | KIAA1470 | MACF2 | dystonin | - | HPRD,BioGRID | 14581450 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-Western | BioGRID | 14600259 |
| DYNC1I2 | DNCI2 | FLJ21089 | FLJ90842 | IC2 | MGC104199 | MGC111094 | MGC9324 | dynein, cytoplasmic 1, intermediate chain 2 | - | HPRD | 8522607 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | Affinity Capture-Western | BioGRID | 10189366 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 8955163 |
| GSTK1 | GST13 | glutathione S-transferase kappa 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | - | HPRD,BioGRID | 9361024 |9454836 |
| HTT | HD | IT15 | huntingtin | - | HPRD | 9454836 |
| KIF11 | EG5 | HKSP | KNSL1 | TRIP5 | kinesin family member 11 | - | HPRD,BioGRID | 9235942 |
| MAP2K3 | MAPKK3 | MEK3 | MKK3 | PRKMK3 | mitogen-activated protein kinase kinase 3 | - | HPRD | 15375157 |
| MAP2K6 | MAPKK6 | MEK6 | MKK6 | PRKMK6 | SAPKK3 | mitogen-activated protein kinase kinase 6 | - | HPRD | 15375157 |
| MAPRE1 | EB1 | MGC117374 | MGC129946 | microtubule-associated protein, RP/EB family, member 1 | - | HPRD | 12388762 |
| PAFAH1B1 | LIS1 | LIS2 | MDCR | MDS | PAFAH | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | - | HPRD,BioGRID | 11889140|14584027 |
| PLK1 | PLK | STPK13 | polo-like kinase 1 (Drosophila) | Affinity Capture-Western | BioGRID | 12852857 |
| RAB6A | RAB6 | RAB6A' | RAB6B | RAB6A, member RAS oncogene family | Reconstituted Complex Two-hybrid | BioGRID | 12401177 |
| RFXANK | ANKRA1 | BLS | F14150_1 | MGC138628 | RFX-B | regulatory factor X-associated ankyrin-containing protein | Affinity Capture-MS | BioGRID | 17353931 |
| SAR1A | SAR1 | SARA1 | Sara | masra2 | SAR1 homolog A (S. cerevisiae) | Sar1p (SARA1) interacts with an unspecified isoform of p150glued (DCTN1). This interaction is probably indirect and bridged by Sec23p-Sec24p. | BIND | 15580264 |
| SEC23A | CLSD | MGC26267 | Sec23 homolog A (S. cerevisiae) | Sec23A interacts with p150glued (DCTN1). | BIND | 15580264 |
| SEC24D | FLJ43974 | KIAA0755 | SEC24 family, member D (S. cerevisiae) | Sec24D interacts with p150glued (DCTN1). | BIND | 15580264 |
| SPTBN1 | ELF | SPTB2 | betaSpII | spectrin, beta, non-erythrocytic 1 | Affinity Capture-Western | BioGRID | 8991093 |
| SPTBN2 | GTRAP41 | SCA5 | spectrin, beta, non-erythrocytic 2 | - | HPRD,BioGRID | 11461920 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-Western | BioGRID | 15611139 |
| VIM | FLJ36605 | vimentin | - | HPRD,BioGRID | 14600259 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | 46 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA UP | 22 | 12 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| JAIN NFKB SIGNALING | 75 | 44 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| MIZUSHIMA AUTOPHAGOSOME FORMATION | 19 | 15 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 240 | 246 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.