Gene Page: DEFB1
Summary ?
| GeneID | 1672 |
| Symbol | DEFB1 |
| Synonyms | BD1|DEFB-1|DEFB101|HBD1 |
| Description | defensin beta 1 |
| Reference | MIM:602056|HGNC:HGNC:2766|Ensembl:ENSG00000164825|HPRD:03634|Vega:OTTHUMG00000090367 |
| Gene type | protein-coding |
| Map location | 8p23.1 |
| Pascal p-value | 0.161 |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1320385 | chr1 | 54064468 | DEFB1 | 1672 | 0.14 | trans | ||
| rs12036675 | chr1 | 54066317 | DEFB1 | 1672 | 0.14 | trans | ||
| rs17131472 | chr1 | 91962966 | DEFB1 | 1672 | 0.02 | trans | ||
| rs2976809 | chr3 | 125451738 | DEFB1 | 1672 | 0.06 | trans | ||
| rs7448349 | 0 | DEFB1 | 1672 | 0.07 | trans | |||
| rs10050805 | chr5 | 30805352 | DEFB1 | 1672 | 0.14 | trans | ||
| rs9381511 | chr6 | 12931447 | DEFB1 | 1672 | 0.1 | trans | ||
| rs819517 | chr6 | 47052415 | DEFB1 | 1672 | 0.14 | trans | ||
| rs10948326 | chr6 | 47062800 | DEFB1 | 1672 | 0.14 | trans | ||
| rs1159612 | chr10 | 85329057 | DEFB1 | 1672 | 0.05 | trans | ||
| rs10879027 | chr12 | 70237156 | DEFB1 | 1672 | 0.11 | trans | ||
| rs16958302 | chr16 | 57647432 | DEFB1 | 1672 | 0.16 | trans | ||
| rs6095741 | chr20 | 48666589 | DEFB1 | 1672 | 3.2E-6 | trans | ||
| rs5915713 | chrX | 4077883 | DEFB1 | 1672 | 0.02 | trans | ||
| rs5991662 | chrX | 43335796 | DEFB1 | 1672 | 0.01 | trans | ||
| rs1545676 | chrX | 93530472 | DEFB1 | 1672 | 0.01 | trans |
Section II. Transcriptome annotation
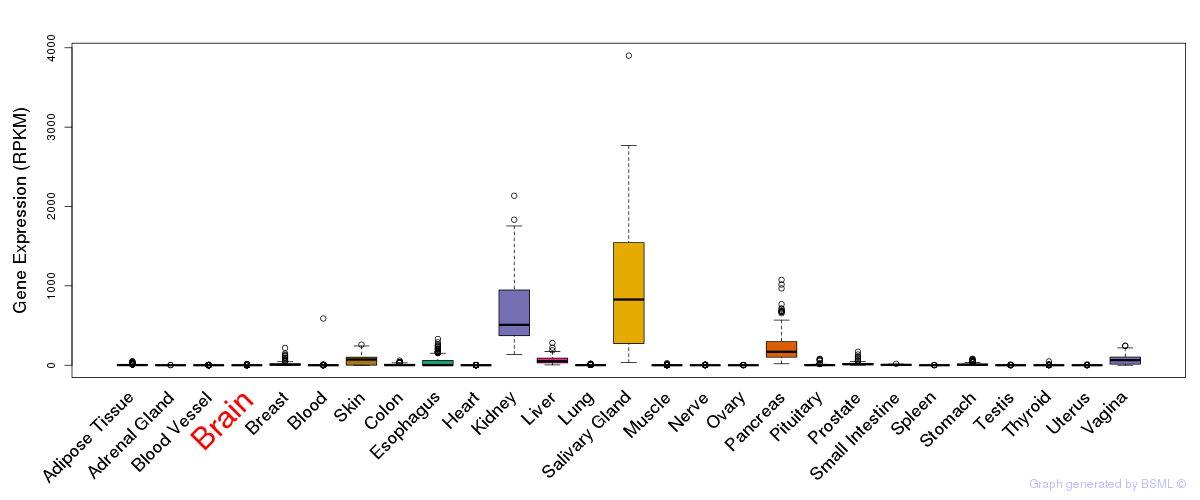
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEFENSINS | 51 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME BETA DEFENSINS | 42 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 BULK DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| SPIRA SMOKERS LUNG CANCER UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |