Gene Page: DFNA5
Summary ?
| GeneID | 1687 |
| Symbol | DFNA5 |
| Synonyms | ICERE-1 |
| Description | deafness, autosomal dominant 5 |
| Reference | MIM:608798|HGNC:HGNC:2810|Ensembl:ENSG00000105928|HPRD:02996|Vega:OTTHUMG00000023237 |
| Gene type | protein-coding |
| Map location | 7p15 |
| Pascal p-value | 1.195E-4 |
| Sherlock p-value | 0.626 |
| Fetal beta | 0.787 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11645781 | chr16 | 6896127 | DFNA5 | 1687 | 0.16 | trans | ||
| rs1001821 | 7 | 24796541 | DFNA5 | ENSG00000105928.9 | 6.942E-7 | 0.04 | 12703 | gtex_brain_ba24 |
| rs138304920 | 7 | 24548616 | DFNA5 | ENSG00000105928.9 | 1.28E-7 | 0 | 260628 | gtex_brain_putamen_basal |
| rs141777824 | 7 | 24548617 | DFNA5 | ENSG00000105928.9 | 1.597E-7 | 0 | 260627 | gtex_brain_putamen_basal |
| rs11770891 | 7 | 24554565 | DFNA5 | ENSG00000105928.9 | 5.671E-9 | 0 | 254679 | gtex_brain_putamen_basal |
| rs2255365 | 7 | 24561661 | DFNA5 | ENSG00000105928.9 | 9.235E-9 | 0 | 247583 | gtex_brain_putamen_basal |
| rs2711103 | 7 | 24563064 | DFNA5 | ENSG00000105928.9 | 9.371E-9 | 0 | 246180 | gtex_brain_putamen_basal |
| rs2158890 | 7 | 24564143 | DFNA5 | ENSG00000105928.9 | 8.566E-9 | 0 | 245101 | gtex_brain_putamen_basal |
| rs2529075 | 7 | 24569527 | DFNA5 | ENSG00000105928.9 | 7.389E-9 | 0 | 239717 | gtex_brain_putamen_basal |
| rs2529042 | 7 | 24607175 | DFNA5 | ENSG00000105928.9 | 7.415E-12 | 0 | 202069 | gtex_brain_putamen_basal |
| rs2108301 | 7 | 24660345 | DFNA5 | ENSG00000105928.9 | 6.143E-10 | 0 | 148899 | gtex_brain_putamen_basal |
| rs2721791 | 7 | 24680389 | DFNA5 | ENSG00000105928.9 | 9.043E-12 | 0 | 128855 | gtex_brain_putamen_basal |
| rs71537294 | 7 | 24680394 | DFNA5 | ENSG00000105928.9 | 1.5E-10 | 0 | 128850 | gtex_brain_putamen_basal |
| rs2711133 | 7 | 24680402 | DFNA5 | ENSG00000105928.9 | 6.317E-11 | 0 | 128842 | gtex_brain_putamen_basal |
| rs2529083 | 7 | 24702513 | DFNA5 | ENSG00000105928.9 | 5.208E-10 | 0 | 106731 | gtex_brain_putamen_basal |
| rs722218 | 7 | 24718710 | DFNA5 | ENSG00000105928.9 | 6.016E-10 | 0 | 90534 | gtex_brain_putamen_basal |
| rs2529037 | 7 | 24722178 | DFNA5 | ENSG00000105928.9 | 5.714E-10 | 0 | 87066 | gtex_brain_putamen_basal |
| rs2721808 | 7 | 24744309 | DFNA5 | ENSG00000105928.9 | 7.693E-10 | 0 | 64935 | gtex_brain_putamen_basal |
| rs2721809 | 7 | 24747910 | DFNA5 | ENSG00000105928.9 | 2.923E-10 | 0 | 61334 | gtex_brain_putamen_basal |
| rs2721810 | 7 | 24748565 | DFNA5 | ENSG00000105928.9 | 1.011E-9 | 0 | 60679 | gtex_brain_putamen_basal |
| rs2521755 | 7 | 24749223 | DFNA5 | ENSG00000105928.9 | 7.693E-10 | 0 | 60021 | gtex_brain_putamen_basal |
| rs2721811 | 7 | 24749429 | DFNA5 | ENSG00000105928.9 | 8.103E-11 | 0 | 59815 | gtex_brain_putamen_basal |
| rs1544366 | 7 | 24758248 | DFNA5 | ENSG00000105928.9 | 7.303E-11 | 0 | 50996 | gtex_brain_putamen_basal |
| rs398094926 | 7 | 24758438 | DFNA5 | ENSG00000105928.9 | 7.693E-10 | 0 | 50806 | gtex_brain_putamen_basal |
| rs876308 | 7 | 24758565 | DFNA5 | ENSG00000105928.9 | 8.103E-11 | 0 | 50679 | gtex_brain_putamen_basal |
| rs876307 | 7 | 24758645 | DFNA5 | ENSG00000105928.9 | 8.103E-11 | 0 | 50599 | gtex_brain_putamen_basal |
| rs876305 | 7 | 24758795 | DFNA5 | ENSG00000105928.9 | 8.103E-11 | 0 | 50449 | gtex_brain_putamen_basal |
| rs2023793 | 7 | 24759063 | DFNA5 | ENSG00000105928.9 | 8.103E-11 | 0 | 50181 | gtex_brain_putamen_basal |
| rs2521757 | 7 | 24761669 | DFNA5 | ENSG00000105928.9 | 7.693E-10 | 0 | 47575 | gtex_brain_putamen_basal |
| rs2521758 | 7 | 24762292 | DFNA5 | ENSG00000105928.9 | 7.693E-10 | 0 | 46952 | gtex_brain_putamen_basal |
| rs2521759 | 7 | 24764655 | DFNA5 | ENSG00000105928.9 | 8.484E-10 | 0 | 44589 | gtex_brain_putamen_basal |
| rs2721795 | 7 | 24766614 | DFNA5 | ENSG00000105928.9 | 7.288E-11 | 0 | 42630 | gtex_brain_putamen_basal |
| rs2521765 | 7 | 24772641 | DFNA5 | ENSG00000105928.9 | 2.688E-9 | 0 | 36603 | gtex_brain_putamen_basal |
| rs2721805 | 7 | 24780316 | DFNA5 | ENSG00000105928.9 | 2.503E-9 | 0 | 28928 | gtex_brain_putamen_basal |
| rs2721803 | 7 | 24781532 | DFNA5 | ENSG00000105928.9 | 6.667E-10 | 0 | 27712 | gtex_brain_putamen_basal |
| rs2521768 | 7 | 24784403 | DFNA5 | ENSG00000105928.9 | 1.23E-6 | 0 | 24841 | gtex_brain_putamen_basal |
| rs2521770 | 7 | 24789082 | DFNA5 | ENSG00000105928.9 | 4.569E-9 | 0 | 20162 | gtex_brain_putamen_basal |
| rs1476520 | 7 | 24796229 | DFNA5 | ENSG00000105928.9 | 1.152E-8 | 0 | 13015 | gtex_brain_putamen_basal |
| rs1476522 | 7 | 24796693 | DFNA5 | ENSG00000105928.9 | 2.408E-8 | 0 | 12551 | gtex_brain_putamen_basal |
| rs2521772 | 7 | 24799496 | DFNA5 | ENSG00000105928.9 | 1.151E-9 | 0 | 9748 | gtex_brain_putamen_basal |
| rs2521773 | 7 | 24801999 | DFNA5 | ENSG00000105928.9 | 9.612E-11 | 0 | 7245 | gtex_brain_putamen_basal |
| rs6976667 | 7 | 24805050 | DFNA5 | ENSG00000105928.9 | 2.61E-7 | 0 | 4194 | gtex_brain_putamen_basal |
| rs397818230 | 7 | 24807112 | DFNA5 | ENSG00000105928.9 | 6.325E-7 | 0 | 2132 | gtex_brain_putamen_basal |
| rs4722389 | 7 | 24807995 | DFNA5 | ENSG00000105928.9 | 3.392E-7 | 0 | 1249 | gtex_brain_putamen_basal |
| rs7787684 | 7 | 24820215 | DFNA5 | ENSG00000105928.9 | 1.008E-11 | 0 | -10971 | gtex_brain_putamen_basal |
| rs4719781 | 7 | 24832308 | DFNA5 | ENSG00000105928.9 | 6.406E-12 | 0 | -23064 | gtex_brain_putamen_basal |
| rs2106760 | 7 | 24835787 | DFNA5 | ENSG00000105928.9 | 2.675E-8 | 0 | -26543 | gtex_brain_putamen_basal |
| rs10227466 | 7 | 24843621 | DFNA5 | ENSG00000105928.9 | 3.054E-9 | 0 | -34377 | gtex_brain_putamen_basal |
| rs739627 | 7 | 24853496 | DFNA5 | ENSG00000105928.9 | 6.362E-7 | 0 | -44252 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
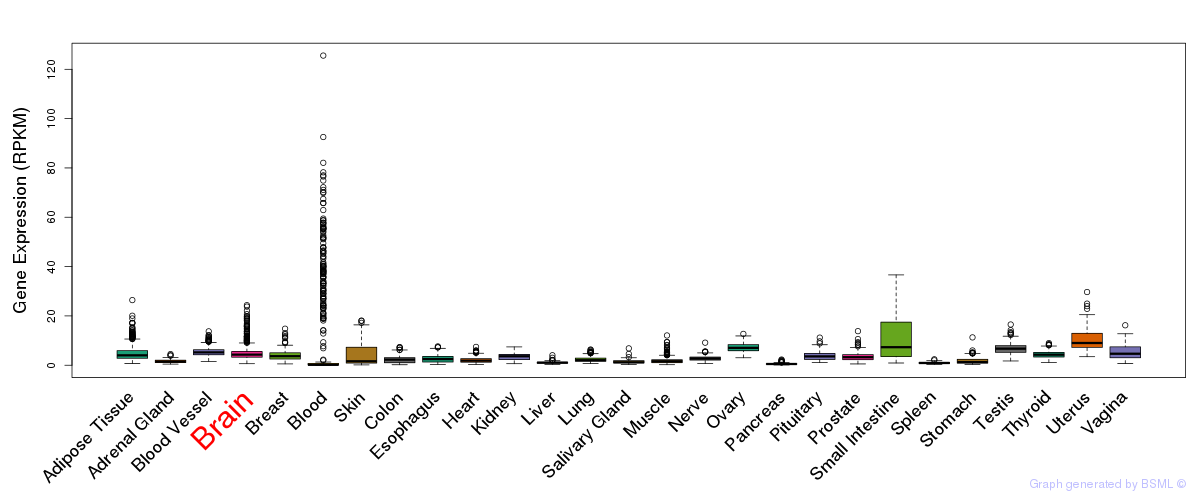
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL UP | 63 | 39 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1 | 45 | 27 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS UP | 42 | 26 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN | 78 | 49 | All SZGR 2.0 genes in this pathway |
| CUI GLUCOSE DEPRIVATION | 60 | 44 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE UP | 40 | 23 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR UP | 33 | 25 | All SZGR 2.0 genes in this pathway |