Gene Page: DLX3
Summary ?
| GeneID | 1747 |
| Symbol | DLX3 |
| Synonyms | AI4|TDO |
| Description | distal-less homeobox 3 |
| Reference | MIM:600525|HGNC:HGNC:2916|Ensembl:ENSG00000064195|HPRD:02752|Vega:OTTHUMG00000161892 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.954 |
| Fetal beta | -0.257 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05639912 | 17 | 48070891 | DLX3 | 3.562E-4 | -0.51 | 0.042 | DMG:Wockner_2014 |
| cg24383056 | 17 | 48071706 | DLX3 | 4.376E-4 | -0.282 | 0.045 | DMG:Wockner_2014 |
| cg07394955 | 17 | 48070699 | DLX3 | 4.82E-4 | -0.365 | 0.046 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
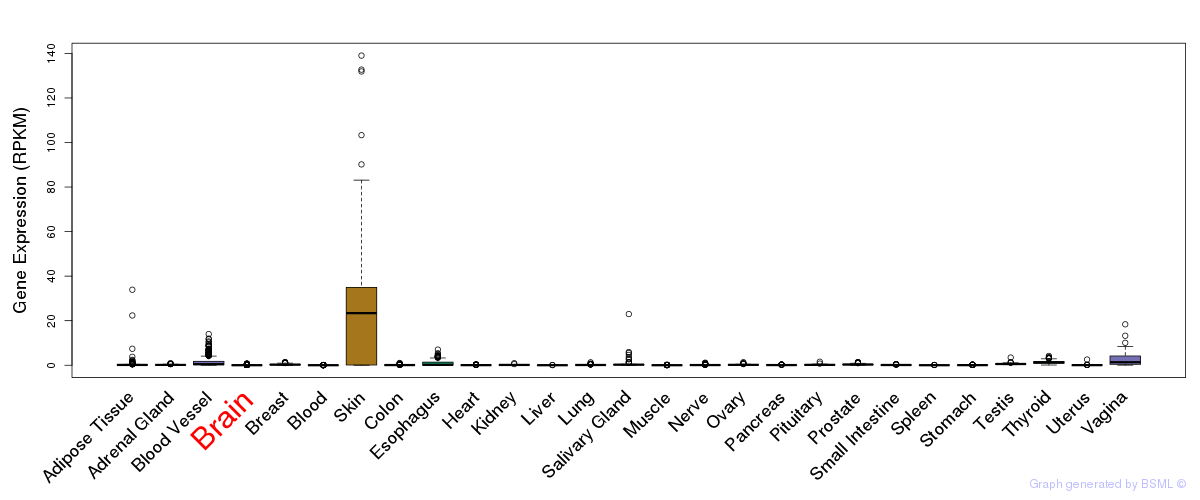
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MICAL2 | 0.94 | 0.87 |
| CNTNAP1 | 0.93 | 0.88 |
| GRIN1 | 0.92 | 0.94 |
| PACSIN1 | 0.92 | 0.81 |
| PTPRN | 0.92 | 0.94 |
| MFSD4 | 0.92 | 0.87 |
| TYRO3 | 0.91 | 0.88 |
| CAMTA2 | 0.91 | 0.92 |
| SYNGR1 | 0.91 | 0.91 |
| AC105206.1 | 0.91 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.52 | -0.57 |
| C9orf46 | -0.49 | -0.48 |
| GTF3C6 | -0.48 | -0.43 |
| EXOSC8 | -0.45 | -0.37 |
| C21orf57 | -0.45 | -0.42 |
| RPS20 | -0.45 | -0.54 |
| RBMX2 | -0.45 | -0.45 |
| RPL23A | -0.44 | -0.46 |
| RPS19P3 | -0.44 | -0.51 |
| DYNLT1 | -0.44 | -0.49 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| ZAIDI OSTEOBLAST TRANSCRIPTION FACTORS | 14 | 12 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |