Gene Page: DMRT1
Summary ?
| GeneID | 1761 |
| Symbol | DMRT1 |
| Synonyms | CT154|DMT1 |
| Description | doublesex and mab-3 related transcription factor 1 |
| Reference | MIM:602424|HGNC:HGNC:2934|Ensembl:ENSG00000137090|HPRD:03885|Vega:OTTHUMG00000019435 |
| Gene type | protein-coding |
| Map location | 9p24.3 |
| Pascal p-value | 0.058 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
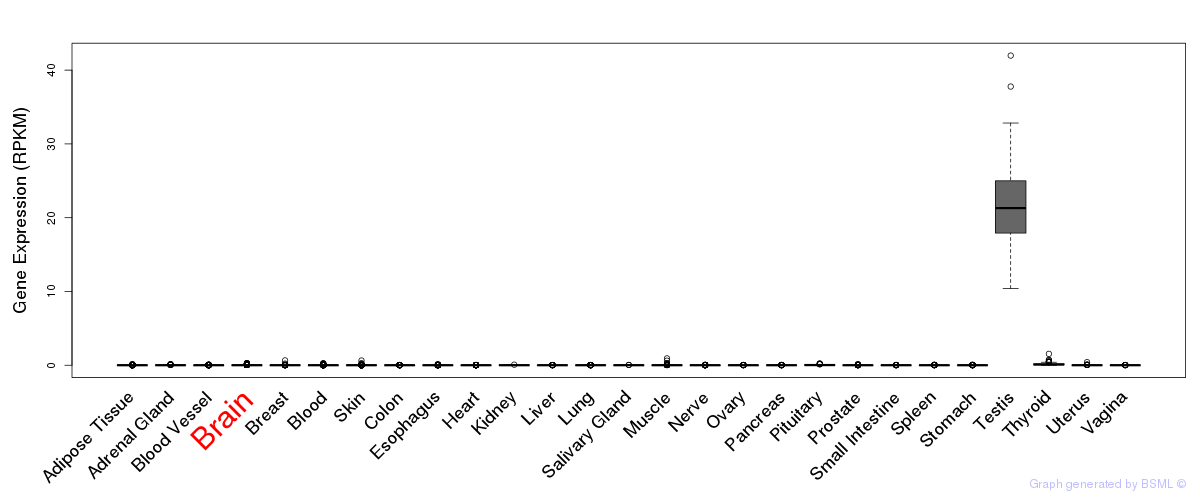
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF512B | 0.87 | 0.88 |
| ZNF785 | 0.86 | 0.88 |
| ZNF710 | 0.86 | 0.83 |
| ARHGEF10L | 0.86 | 0.87 |
| ZSCAN22 | 0.86 | 0.84 |
| SLC35E2 | 0.86 | 0.85 |
| DOT1L | 0.86 | 0.88 |
| TNRC18B | 0.86 | 0.85 |
| RAI1 | 0.86 | 0.84 |
| KLHL17 | 0.85 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.60 | -0.72 |
| AF347015.27 | -0.59 | -0.72 |
| MT-CO2 | -0.57 | -0.70 |
| COPZ2 | -0.57 | -0.67 |
| AF347015.33 | -0.56 | -0.66 |
| MT-CYB | -0.56 | -0.67 |
| C5orf53 | -0.55 | -0.58 |
| AF347015.8 | -0.55 | -0.69 |
| B2M | -0.54 | -0.62 |
| HIGD1B | -0.54 | -0.68 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TAKADA GASTRIC CANCER COPY NUMBER DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED HCP IN SPERM DN | 28 | 13 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K27ME3 | 79 | 59 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |