Gene Page: EEF1A1
Summary ?
| GeneID | 1915 |
| Symbol | EEF1A1 |
| Synonyms | CCS-3|CCS3|EE1A1|EEF-1|EEF1A|EF-Tu|EF1A|GRAF-1EF|HNGC:16303|LENG7|PTI1|eEF1A-1 |
| Description | eukaryotic translation elongation factor 1 alpha 1 |
| Reference | MIM:130590|HGNC:HGNC:3189|Ensembl:ENSG00000156508|HPRD:00559| |
| Gene type | protein-coding |
| Map location | 6q14.1 |
| Pascal p-value | 0.85 |
| Sherlock p-value | 0.003 |
| Fetal beta | 0.715 |
| Support | RNA AND PROTEIN SYNTHESIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2361 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
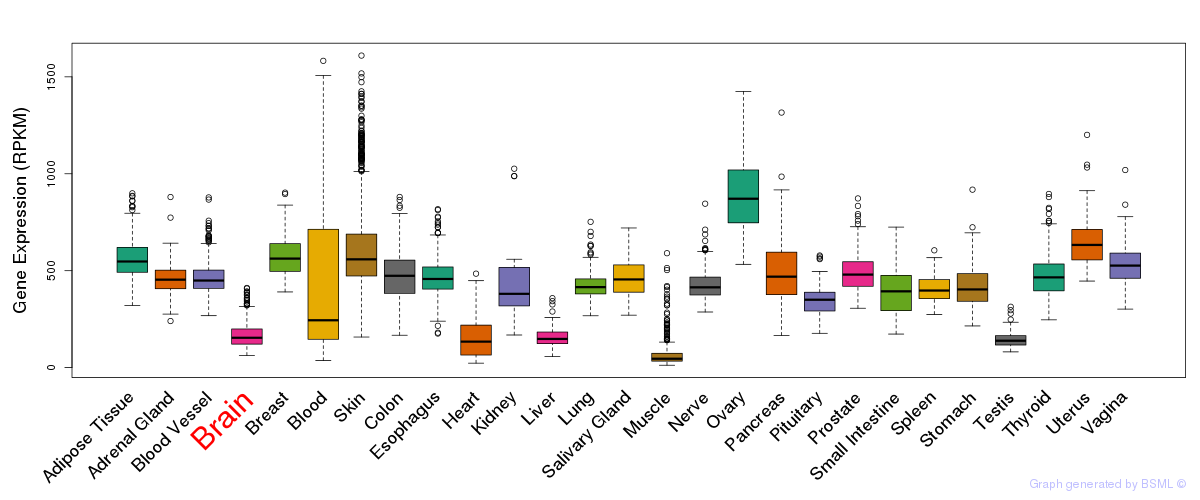
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003746 | translation elongation factor activity | IEA | - | |
| GO:0003746 | translation elongation factor activity | NAS | 3570288 | |
| GO:0003924 | GTPase activity | IEA | - | |
| GO:0003924 | GTPase activity | NAS | 3570288 | |
| GO:0005515 | protein binding | IPI | 15231747 |17373842 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005525 | GTP binding | TAS | 3512269 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006414 | translational elongation | EXP | 15189156 | |
| GO:0006414 | translational elongation | IEA | - | |
| GO:0006414 | translational elongation | NAS | 3570288 | |
| GO:0006414 | translational elongation | TAS | 8812466 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10368288 | |
| GO:0005853 | eukaryotic translation elongation factor 1 complex | TAS | 2564392 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005737 | cytoplasm | NAS | - | |
| GO:0005737 | cytoplasm | TAS | 3512269 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANKRD24 | KIAA1981 | ankyrin repeat domain 24 | Two-hybrid | BioGRID | 16169070 |
| ARIH2 | ARI2 | FLJ10938 | FLJ33921 | TRIAD1 | ariadne homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| AXIN1 | AXIN | MGC52315 | axin 1 | Two-hybrid | BioGRID | 16169070 |
| BRMS1 | DKFZp564A063 | breast cancer metastasis suppressor 1 | Two-hybrid | BioGRID | 16169070 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| C11orf59 | FLJ20625 | chromosome 11 open reading frame 59 | Two-hybrid | BioGRID | 16169070 |
| C11orf79 | FLJ20487 | chromosome 11 open reading frame 79 | Two-hybrid | BioGRID | 16169070 |
| C4B | C4A | C4A13 | C4A91 | C4B1 | C4B12 | C4B2 | C4B3 | C4B5 | C4F | CH | CO4 | CPAMD3 | FLJ60561 | MGC164979 | complement component 4B (Childo blood group) | - | HPRD,BioGRID | 10851272 |
| CCDC130 | MGC10471 | coiled-coil domain containing 130 | Two-hybrid | BioGRID | 16169070 |
| CCL18 | AMAC-1 | AMAC1 | CKb7 | DC-CK1 | DCCK1 | MIP-4 | PARC | SCYA18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | Two-hybrid | BioGRID | 16169070 |
| CKS2 | CKSHS2 | CDC28 protein kinase regulatory subunit 2 | Two-hybrid | BioGRID | 16169070 |
| COX17 | MGC104397 | MGC117386 | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| CRADD | MGC9163 | RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain | Two-hybrid | BioGRID | 16169070 |
| CRCT1 | C1orf42 | NICE-1 | cysteine-rich C-terminal 1 | Two-hybrid | BioGRID | 16169070 |
| DARS | DKFZp781B11202 | MGC111579 | aspartyl-tRNA synthetase | - | HPRD,BioGRID | 7806521 |
| DLEU1 | BCMS | DLB1 | DLEU2 | LEU1 | LEU2 | MGC22430 | NCRNA00021 | XTP6 | deleted in lymphocytic leukemia 1 (non-protein coding) | Two-hybrid | BioGRID | 16169070 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Two-hybrid | BioGRID | 16169070 |
| EIF3F | EIF3S5 | eIF3-p47 | eukaryotic translation initiation factor 3, subunit F | Two-hybrid | BioGRID | 16169070 |
| EXOSC4 | FLJ20591 | RRP41 | RRP41A | Rrp41p | SKI6 | Ski6p | hRrp41p | p12A | exosome component 4 | - | HPRD,BioGRID | 15231747 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Two-hybrid | BioGRID | 16169070 |
| HBXIP | MGC71071 | XIP | hepatitis B virus x interacting protein | Two-hybrid | BioGRID | 16169070 |
| HSPE1 | CPN10 | GROES | HSP10 | heat shock 10kDa protein 1 (chaperonin 10) | Two-hybrid | BioGRID | 16169070 |
| ITSN1 | ITSN | MGC134948 | MGC134949 | SH3D1A | SH3P17 | intersectin 1 (SH3 domain protein) | Two-hybrid | BioGRID | 16169070 |
| KIF1B | CMT2 | CMT2A | CMT2A1 | FLJ23699 | HMSNII | KIAA0591 | KIAA1448 | KLP | MGC134844 | kinesin family member 1B | Two-hybrid | BioGRID | 16169070 |
| LAMA4 | DKFZp686D23145 | LAMA3 | LAMA4*-1 | laminin, alpha 4 | Two-hybrid | BioGRID | 16169070 |
| LSM3 | SMX4 | USS2 | YLR438C | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | Two-hybrid | BioGRID | 16169070 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 14743216 |
| MLLT3 | AF9 | FLJ2035 | YEATS3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | Two-hybrid | BioGRID | 16169070 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | Two-hybrid | BioGRID | 16169070 |
| MRPL42 | HSPC204 | MRP-L31 | MRPL31 | MRPS32 | PTD007 | RPML31 | mitochondrial ribosomal protein L42 | Two-hybrid | BioGRID | 16169070 |
| NEU1 | FLJ93471 | NANH | NEU | SIAL1 | sialidase 1 (lysosomal sialidase) | Two-hybrid | BioGRID | 16169070 |
| ORMDL3 | - | ORM1-like 3 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| PAEP | GD | GdA | GdF | GdS | MGC138509 | MGC142288 | PAEG | PEP | PP14 | progestagen-associated endometrial protein | Two-hybrid | BioGRID | 16169070 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PAPSS1 | ATPSK1 | PAPSS | SK1 | 3'-phosphoadenosine 5'-phosphosulfate synthase 1 | Two-hybrid | BioGRID | 16169070 |
| PCDHA4 | CNR1 | CNRN1 | CRNR1 | MGC138307 | MGC142169 | PCDH-ALPHA4 | protocadherin alpha 4 | Two-hybrid | BioGRID | 16169070 |
| PFN2 | D3S1319E | PFL | profilin 2 | Two-hybrid | BioGRID | 16169070 |
| PHYHIP | DYRK1AP3 | KIAA0273 | PAHX-AP | PAHXAP1 | phytanoyl-CoA 2-hydroxylase interacting protein | Two-hybrid | BioGRID | 16169070 |
| PLAUR | CD87 | UPAR | URKR | plasminogen activator, urokinase receptor | Two-hybrid | BioGRID | 16169070 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 11886851 |
| POLE2 | DPE2 | polymerase (DNA directed), epsilon 2 (p59 subunit) | Two-hybrid | BioGRID | 16169070 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | Two-hybrid | BioGRID | 16169070 |
| PQBP1 | MRX55 | MRXS3 | MRXS8 | NPW38 | RENS1 | SHS | polyglutamine binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| PSG9 | PSG11 | PSGII | pregnancy specific beta-1-glycoprotein 9 | Two-hybrid | BioGRID | 16169070 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| PTPN4 | PTPMEG | PTPMEG1 | protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) | Two-hybrid | BioGRID | 16169070 |
| PTPRCAP | CD45-AP | LPAP | MGC138602 | MGC138603 | protein tyrosine phosphatase, receptor type, C-associated protein | Two-hybrid | BioGRID | 16169070 |
| RAB27A | GS2 | HsT18676 | MGC117246 | RAB27 | RAM | RAB27A, member RAS oncogene family | Two-hybrid | BioGRID | 16169070 |
| RFC5 | MGC1155 | RFC36 | replication factor C (activator 1) 5, 36.5kDa | Two-hybrid | BioGRID | 16169070 |
| RNF10 | KIAA0262 | MGC126758 | MGC126764 | RIE2 | ring finger protein 10 | Two-hybrid | BioGRID | 16169070 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 16169070 |
| RPLP1 | FLJ27448 | MGC5215 | P1 | RPP1 | ribosomal protein, large, P1 | Two-hybrid | BioGRID | 16169070 |
| RSRC1 | BM-011 | MGC12197 | arginine/serine-rich coiled-coil 1 | Two-hybrid | BioGRID | 16169070 |
| SARS2 | FLJ20450 | SARS | SARSM | SERS | SYS | SerRSmt | mtSerRS | seryl-tRNA synthetase 2, mitochondrial | - | HPRD | 15109557 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| SSR1 | DKFZp781N23103 | FLJ14232 | FLJ22100 | FLJ23034 | FLJ78242 | FLJ93042 | TRAPA | signal sequence receptor, alpha | Two-hybrid | BioGRID | 16169070 |
| STMN2 | SCG10 | SCGN10 | SGC10 | stathmin-like 2 | Two-hybrid | BioGRID | 16169070 |
| SULT1E1 | EST | EST-1 | MGC34459 | STE | sulfotransferase family 1E, estrogen-preferring, member 1 | Two-hybrid | BioGRID | 16169070 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | Two-hybrid | BioGRID | 16169070 |
| TGIF1 | HPE4 | MGC39747 | MGC5066 | TGIF | TGFB-induced factor homeobox 1 | Two-hybrid | BioGRID | 16169070 |
| TMPRSS3 | DFNB10 | DFNB8 | ECHOS1 | TADG12 | transmembrane protease, serine 3 | Two-hybrid | BioGRID | 16169070 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | Two-hybrid | BioGRID | 16169070 |
| TPT1 | FLJ27337 | HRF | TCTP | p02 | tumor protein, translationally-controlled 1 | - | HPRD | 14623968 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
| TRDMT1 | DNMT2 | M.HsaIIP | PuMet | RNMT1 | tRNA aspartic acid methyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | Two-hybrid | BioGRID | 16169070 |
| UBQLN4 | A1U | C1orf6 | CIP75 | UBIN | ubiquilin 4 | Two-hybrid | BioGRID | 16169070 |
| VARS | G7A | VARS1 | VARS2 | valyl-tRNA synthetase | - | HPRD,BioGRID | 2556394 |3169261 |
| WARS | GAMMA-2 | IFI53 | IFP53 | tryptophanyl-tRNA synthetase | WRS interacts with EF-1-alpha. | BIND | 11829477 |
| XPO5 | FLJ14239 | FLJ32057 | FLJ45606 | KIAA1291 | exportin 5 | - | HPRD,BioGRID | 12426393 |
| XRN2 | - | 5'-3' exoribonuclease 2 | - | HPRD,BioGRID | 15231747 |
| ZCCHC10 | FLJ20094 | zinc finger, CCHC domain containing 10 | Two-hybrid | BioGRID | 16169070 |
| ZNF24 | KOX17 | RSG-A | ZNF191 | ZSCAN3 | Zfp191 | zinc finger protein 24 | Two-hybrid | BioGRID | 16169070 |
| ZNF259 | MGC110983 | ZPR1 | zinc finger protein 259 | - | HPRD,BioGRID | 9852145 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE CHAIN ELONGATION | 153 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 DN | 67 | 43 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID WITH 7Q DELETION DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING DN | 35 | 24 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION UP | 39 | 29 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| KIM LRRC3B TARGETS | 30 | 24 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO PROXIMAL DENDRITES | 37 | 26 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| IRITANI MAD1 TARGETS DN | 47 | 30 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM AND RAPAMYCIN SENSITIVE GENES | 68 | 35 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM RESPONSE TRANSLATION | 37 | 20 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-33 | 116 | 122 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-382 | 235 | 241 | m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-542-3p | 253 | 259 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-543 | 148 | 154 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.